Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- The Global Impact Factor (GIF)
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
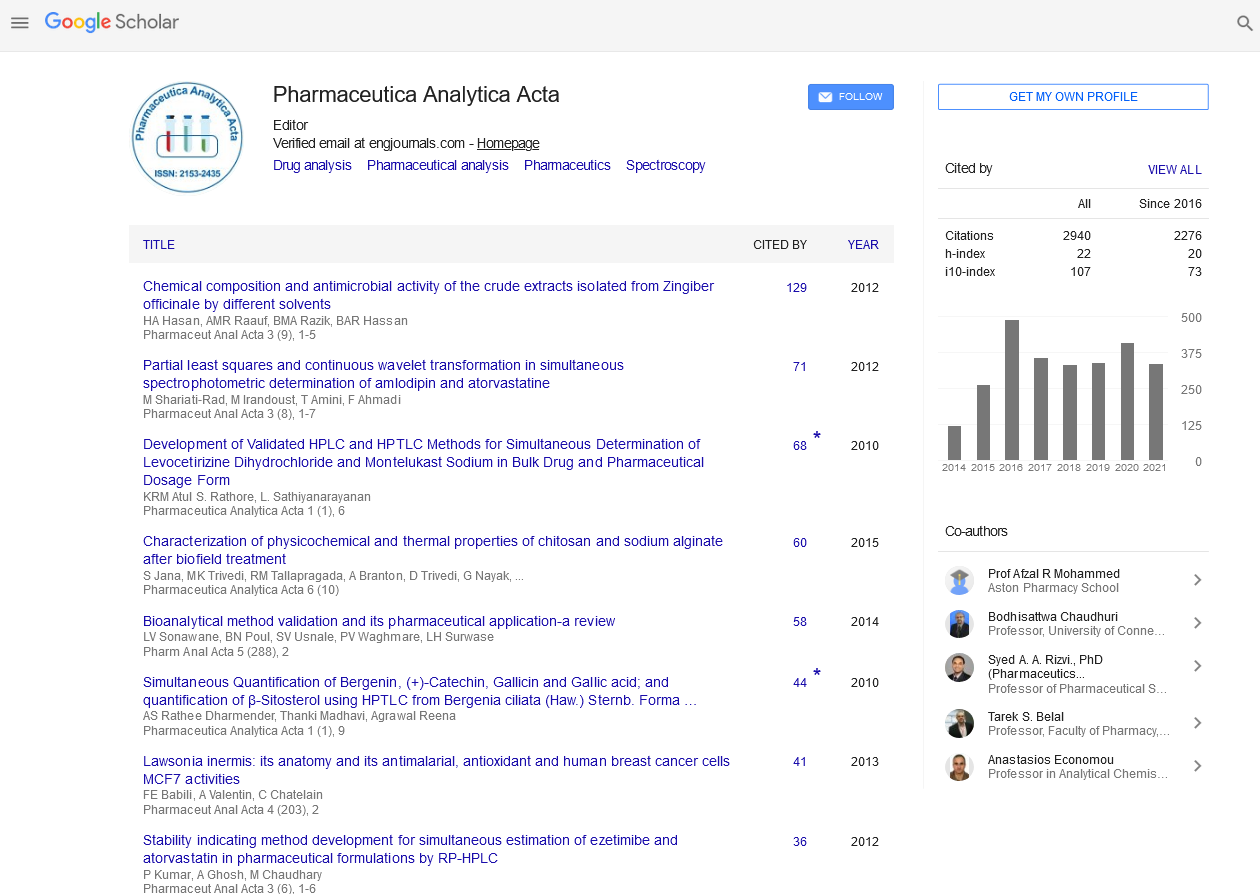
Affinity Determinations of Therapeutic Antibodies to Soluble Native Antigens in Human Serum
2nd International Conference on Pharmaceutics & Novel Drug Delivery Systems
20-22 February 2012 San Francisco Airport Marriott Waterfront, USA
Christine Bee
Scientific Tracks Abstracts: Pharm Anal Acta
Abstract:
Therapeutic antibodies are engineered or selected to have high on-target binding affinities, with equilibrium dissociation constants (KD values) often in the picomolar or subpicomolar range. Characterizing these tight interactions with label-free biosensors, however, can be challenging. We explored the dynamic range of the Kinetic Exclusion Assay (KinExA) in determining solution affinities of antigen/antibody interactions that spanned a 500,000-fold range in their apparent KD values, from 300 fM to 150 nM. These affinity measurements were in excellent agreement with those obtained on Biacore for interactions that fell within the dynamic range of both methods (i.e., KD values of single digit pM and higher). By tailoring the KinExA to each studied antigen, we were not only able to obtain KD values for unpurified recombinant and native antigens contained in conditioned media but extended the application to determining the affinity of therapeutic antibodies towards their native target contained in human serum. We present methods to obtain both the affinity of an interaction and the active concentration of one binding partner relative to another in a single experiment, which increases the information content of an experiment and reduces the consumption of precious serum samples.
Biography :
Christine Bee graduated from Ruhr-University Bochum in Germany with a doctoral degree in biochemistry in which she studied a proapoptotic, multidomain Ras effector protein. Since 2004, Christine has specialized in the biophysical characterization of protein interactions using different fluorescence-based and label-free methods. She was awarded two fellowships from the German National Academic Foundation. Christine now works as a Senior Scientist in the protein engineering department at Rinat-Pfizer in South San Francisco where she uses a variety of biosensor technologies to support the discovery of therapeutic antibodies, especially in lead optimizations..