Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2021) Volume 12, Issue 8
The Intriguing Extrapolations of Haemolysis Assay as Screening Criterion for Selecting Biosurfactant- Producing Microorganisms in Petroleum Industries Process-Conditions
Adenike AOO1,2,4* and Falode OA1,32Department of Microbiology, Faculty of Science, University of Ibadan, Oyo State, Nigeria
3Department of Petroleum Engineering, Faculty of Technology, University of Ibadan, Oyo State, Nigeria
4Pegasus-Zion Community and Environmental Health, Nigeria
Received: 10-Jul-2021 Published: 21-Sep-2021, DOI: 10.35248/2157-7463.21.12.431
Abstract
Commonly-used screening techniques for determination of biosurfactants production by microorganisms include haemolysis assay, generally depicted to confirm the ability of microorganisms in production of biosurfactants. Diameters of zones of haemolysis surrounding microbial colonies are considered as quantitative indication of biosurfactant production whereas; haemolytic reactions on blood agar plates are specifically associated with pathologic types of erythrocyte lysis by microorganisms, due to haemolysins production. Haemolytic microorganisms can destroy erythrocyte membranes, by compromise in integrity of cytoplasmic membranes, through pore-forming mechanisms, multiple-hit mechanism, formation of sphaerocytes, derangement of membrane integrity, detergentlike action, or lipase activity. Relative levels of acute toxicity, cell invasiveness and virulence factors, which can make biosurfactants become opportunistic pathogens that use haemin or haemoglobin as a source of iron, have also been reported. Haemolysins are further classically defined as exotoxins that can be thermostable, and can cross membranes of microorganisms. Haemolysis assay thus, identifies haemolytic microbial strains with lytic, pathogenic, toxigenic, and/or virulent potentials, rather than biosurfactant-producing potential, as the assay does not correlate particularly with specific characteristics of biosurfactants’ production. However, based on new insights and perspectives appropriately extrapolated for the first time in this report, microbial haemolysis assay is considered, the easiest, most-economical, non-animal-based, highly-determinative, reliable and sensitive biosafety selection criterion protocol, for selection of safe and environmental-friendly biosurfactant candidates, for the petroleum industries’ process conditions.
Keywords
Biosurfactants; Blood agar; Microbial haemolysis assay; Microbial toxicity; Petroleum industries
Introduction
Microbial surfactants (biosurfactants) are one of the wide ranges of extracellular compounds that are produced by microorganisms, particularly, bacteria and fungi (yeasts, moulds and mushroom), more especially when grown on hydrophobic substrates. They are surface-active microbial amphiphilic compounds, which are produced on living surfaces, mostly, on microbial cell surfaces or excreted as extracellular hydrophobic and hydrophilic moieties. These characteristics thus, confer on the biosurfactants-producing microorganisms, the ability to accumulate between fluid phases, and also possess the characteristic property for reducing surface and interfacial tension, at surfaces and interfaces respectively. By accumulating at the interfaces of immiscible fluids, biosurfactants have been reported to be able to increase the solubility, bioavailability and subsequent biodegradation of hydrophobic or insoluble organic compounds [1-10]. However, biosurfactants use similar mechanisms to the chemical surfactants but mostly with certain more established advantages [11-13].
In addition to having many unique properties and applications, the ability to exhibit biosensoactives (surface‐ active) properties, which lower the surface tension and the interfacial tension of their growth media, allow biosurfactants to play diverse key beneficial roles [13-28]. Furthermore, the vast structural diversities that characterise biosurfactants, may also explain the reason for their continual intrigue to scientific interests [16,29,30]. Increased environmental awareness has been the main driver for the search of biosurfactants, as replacement for chemical surfactants [2,13,24,29-34].
Literature Review
Benefits and applications of biosurfactants in petroleum industries
Biosurfactant production is considered one of the key technologies for development in the 21st century, and biosurfactants are widely applicable in almost every area of human endeavours, especially in the field of petroleum technology and processes. Specifically, due to their efficacy as dispersion and remediation agents, biosurfactants have several potential applications across the oilprocessing chains, and in the formulations of petrochemical products, microbial enhanced oil recovery, anti-corrosives; biocides for sulphate-reducing bacteria; emulsification and emulsified fuels; de-emulsification; oil waste treatment; enhancement of crude oil transportation through pipelines; crude oil spill clean-ups/ bioremediation of crude-oil polluted soils; including the removal of crude oil from contaminated soils and water bodies by indigenous microbes, biodegradation. Some other benefits of biosurfactants are environmental remediation processes like- oil storage bottom sludge tank cleaning; sediment remediation, soil washing and soil flushing, extraction of bitumen from tar sands, and extraction of hydrocarbon compounds from oil shales, in order to utilise them as a substitute for petroleum energy fuel [10,12,16,17,22,24,35-86].
Biosurfactants being diverse amphiphilic molecules, with wide structural and functional diversities, and because great diversity also exists a m o n g biosurfactant roducing microorganisms, there is adoption of different screening techniques for their determination; although, almost all the screening methods can give qualitative and/or quantitative results [87-93].
Some screening methods for biosurfactant production, which are basically automated and/or miniaturised rapid-screening assays, are available in present times. However, t h e major regular direct and indirect biosurfactants screening assays are presented in Figure 1 [19,88,58,90,93,94-122].
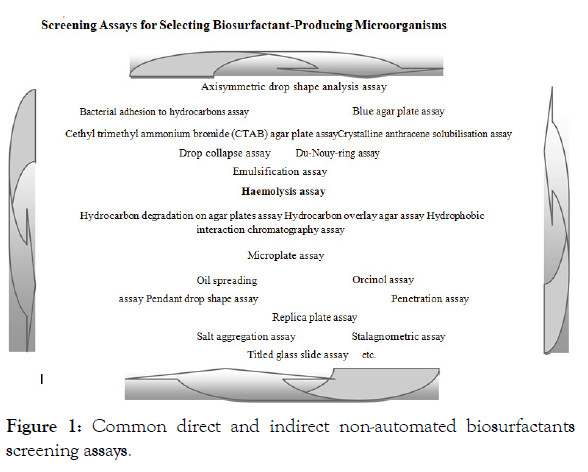
Figure 1: Common direct and indirect non-automated biosurfactants screening assays.
Screening assays for selecting biosurfactant-producing microorganisms
Each biosurfactant screening method, as presented in Figure 1, has its advantages and disadvantages; so, a combination of different methods has been suggested as appropriate for successful screening of biosurfactants [15,87,88,121,123]. In addition to the physiological nature of the biosurfactant-producing microorganisms, screening for biosurfactant producers somehow depends, both on the type of carbon source(s) present, and also the types and amounts of other nutrients in the screening media [92,124-130]. The screening medium used will therefore, tremendously influence production or non-production of biosurfactants; and also influence the type and amount of biosurfactants produced [19].
Haemolysis assay in biosurfactant determination
It was reported that haemolytic activity of biosurfactants was first discovered when Bernheimer and Avigad [131] recorded that surfactin, the biosurfactant produced by B. subtilis, lysed erythrocytes.
Afterwards, haemolysis assay for biosurfactant determination was also reportedly developed by Mulligan et al. [96]. Following the development of haemolysis assay for biosurfactant determination, Carrillo et al. [106] also claimed to have discovered an association between haemolytic activity and surfactant production. Several studies similarly reported the impossibility of biosurfactant production without haemolytic activity, as haemolysis has been referred to as a determination of biosurfactant [87,106,113,121,131-134]. Whereas, as summed up by Kabir et al. [25], very few bacteria would have the selective advantage of lysing erythrocytes, yet haemolysis test has always been considered an ideal assay for determining surfactant production, as it is commonly claimed that biosurfactants cause lysis of erythrocytes, and this is usually the principle adopted in the haemolysis assay for biosurfacts determination.
In several studies on biosurfactants, haemolysis assay on blood agar plates has always been an exclusive experimental screening method to monitor biosurfactant production [19]. Based on the reference that biosurfactant-producing capacity in liquid medium was found to be associated with haemolytic activity, the use of blood agar lysis (haemolysis assay) was considered and recommended as appearing to be a good primary (and in few cases, secondary) screening criterion/method for biosurfactant production, by surfactant-producing microbial strains, and regarded as indicative of biosurfactant production [16,19,25,87,88,96,102,106,113,117,135-145].
Preparation of blood agar for haemolysis assay
Blood agar is an enriched and differential solid growth medium with general composition of-blood agar peptone: 10 g/l; yeast extract: 3 g/l; NaCl: 5 g/l; blood: 100 ml/l (as the basal medium), of which specified mls. of human or animal (rabbit, sheep, horse or cattle) blood is added, for the growth of many microorganisms, especially, the fastidious microorganisms (i.e., microbial species that do not grow easily on general purpose culture media, which are microbial culture media that lack special nutrients). Firstly, for haemolysis assay, the isolated microbial colonies may be subcultured from primary plates, by four-corner streaking or repeated microbial colony transfers (for mostly fungal isolates) on appropriate sterile culture media, in order to obtain pure microbial cultures. The pure microbial cultures can then be inoculated on any of the various modified blood agar plates, such as, Zobell marine medium supplemented with 5% fresh human blood [121]. Other basal culture media to which human or animal blood can be added, in order to prepare blood agar include, blood agar base, tryptone soy agar, nutrient agar, plate count agar, Mueller-Hinton agar, potato dextrose agar, Sabouraud dextrose agar, etc.
Inoculation of blood agar plates is usually followed by incubation at 25-37°C for 24-72°C or 96 hrs, depending on the bacterial or fungal species. The blood agar plates are then visually inspected for haemolysis (clear zone) around the haemolytic microbial colonies. Secondly, the initial isolation of suspected biosurfactantproducers may also primarily be done on blood agar plates, based on the acclaimed ability of many biosurfactants to lyse erythrocytes, which then results in haemolysis around suspected biosurfactant-producing microbial colonies, on the blood agar plates [25,87,131,135,146-148].
Blood agar and haemolysis
Haemolytic microbial strains cause lysis of erythrocytes, and exhibit haemolytic zones, which can be complete or partial haemolysis around the haemolytic microbial colonies. As introduced by Brown, the three basic types of haemolysis (haemolytic reactions) that can be observed on blood agar plates are designated, alpha (∝), beta (β) and gamma (γ) haemolysis [148,149], as denoted in Figure 2.

Figure 2: Haemolytic reactions on blood agar plates.
Alpha-haemolysis (α-haemolysis) is a greenish discoloration that surrounds a haemolytic microbial colony, growing on blood agar plate. This type of haemolysis represents a partial (greenish) lysis or incomplete decomposition (reduction) of the haemoglobin of the erythrocytes (red blood cells). Alpha haemolysis is caused by hydrogen-peroxide produced by alpha-haemolytic bacteria or fungi, which oxidise haemoglobin to green methaemoglobin, in the medium surrounding the colony. Thus, alpha-haemolytic microbes thus, produce greenish diffusible appearance on blood agar plates [149-152].
Beta-haemolysis (β-haemolysis) represents a complete haemolysis (complete breakdown) of the haemoglobin of the red blood cells surrounding a microbial colony, on blood agar plate, giving a transparent or translucent clearing of the blood agar around the microbial colony. Beta haemolysis is more pronounced when the blood agar plate is incubated anaerobically, although some microorganisms are weakly beta-haemolytic species [149].
Gamma-haemolysis (γ-haemolysis/non-haemolysis) is the third type of haemolytic reaction, in which there is actually no haemolysis at all, as there is lack of haemolysis in the area surrounding the microbial colony on blood agar plates. Gamma-haemolysis show neither typical alpha nor beta haemolysis, due to no haemolytic change around the microbial growth on blood agar plates (http://www.encyclopedia.com/science/encyclopedias-almanacs-transcriptsand-maps/blood-agar-hemolysis-and-hemolytic-reactions). There may however, be, slight brownish discolouration (not haemolysis) on the blood agar plates [149]. Zonee of alpha and beta-haemolysis surrounding microbial colonies on blood-agar plates are however, designated as hallmark phenotypic features of various pathogenic microbes.
Haemolysins as microbial toxins and virulence factors
Haemolysins, sometimes classified as enzymes, are lipids and proteins that have been extensively reported and studied in bacteria, fungi, various species of plants, invertebrates, mammals, and also denoted as perforins, in fungi, plants, invertebrates, and mammals [153-168]. Haemolysins cause lysis (destruction) of erythrocytes (red blood cells), by destroying their cell membrane (Figure 3), with release of their haemoglobins; thereby, providing iron, for bacterial growth. Through haemolysins enzymatic attack on phospholipids, the cell membranes are subsequently destabilised [169], as shown in Figure 3.
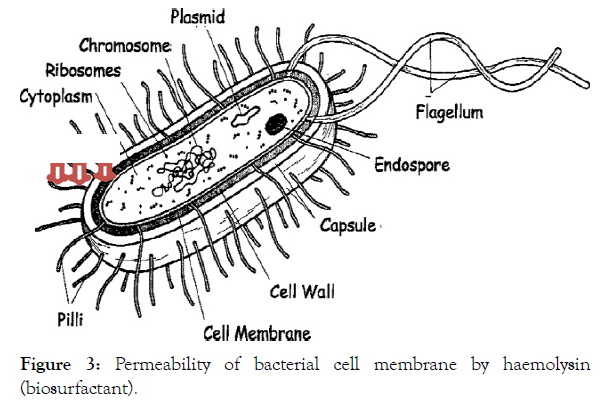
Figure 3: Permeability of bacterial cell membrane by haemolysin (biosurfactant).
Pore formations in microbial cell membranes, derangement of microbial cell membrane integrity, detergent action, or lipase activity are the major mechanisms by which microbial haemolysisns cause haemolysis [69-173]. It was further proposed that the hydrophilic part (the cationic part) of biosurfactants initiate electrostatic interaction with the negatively charged components of the bacterial cell membranes; while the hydrophobic portion was supposed to permit the peptides to insert into, and permeate the bacterial cell membranes [174]. Some haemolysins however, attack the phospholipid of the host cytoplasmic membrane, by using phospholipases lecithinases, and the phospholipids, lecithin (phosphatidylcholine), often used as substrate; although, some haemolysins affect the sterols of the host cytoplasmic membrane [87].
In addition to bacterial growth, due to the release of haemoglobins after red blood lysis; thereby, providing iron, pathogenicity; are also reported, and the responsible haemolysisns considered as toxins. So, being identified as extracellular toxic proteins that are produced by several microbial species, all of which possess a certain pathogenic potential; haemolysins have usually been further considered as virulence factors [175], and sublytic effects of haemolysin can alter host cell regulation and lead to cell death [176,177]. Due to production of cytolytic toxins, haemolysins from several bacterial and fungal strains have been confirmed to possess lytic activities that correlate with severity of haemolysin–induced infections, sometimes, with high mortality rates [168,177-180]. Haemolysis is also considered a pathogenicity indicator tool [180-182], and haemolysins have similarly been linked to increased severity of infections, and concretely associated with virulence, in addition to pathogenesis or pathogenicity [152,183-188].
Among the well-known diverse toxigenic microbial haemolysins are, small β-pore-forming toxins, alpha-haemolysin monomers secreted by Staphylococcus aureus, aerolysin, secreted by Aeromonas hydrophila; α-toxins, secreted by Staphylococcus aureus and Clostridium septicum; cholesterol-dependent cytolysins (CDCs), like streptolysin O, secreted by Streptococcus pyogenes, and listeriolysin O, secreted by Listeria monocytogenes or AB toxins, like the diphtheria toxin, secreted by Corynebacterium diphtheriae, as well as the toxic fungal haemolysins like, nigerlysin, aerolysin, ostreolysin, pleurotolysin A and B etc. [166,169,189-193].
Mechanisms of pore-forming toxins are depicted in Figure 4; thereby, it is designated that pore-forming toxins like thermostable direct haemolysin (TDH) are also known to induce haemolysis, by incorporating into cell membranes to form pores [194]. Pore-forming toxins are secreted by microbial pathogens in a water-soluble form that binds to the target cell, then generally multimerises into an amphipathic structure that finally inserts into the target cell membrane, and then forms a pore [169]. The native thermostable direct haemolysin (TDHn) is transformed into nontoxic fibrils, rich in beta-strands, by incubation at 600°C, to form the incubated thermostable direct haemolysin (TDHi). The TDHi fibrils are dissociated into unfolded states by further heating above 800°C (TDHu) but the protein is trapped in the TDHi structure by slow cooling of TDHu, while rapid cooling of TDHu results in refolding of the protein into toxic TDHn [195].
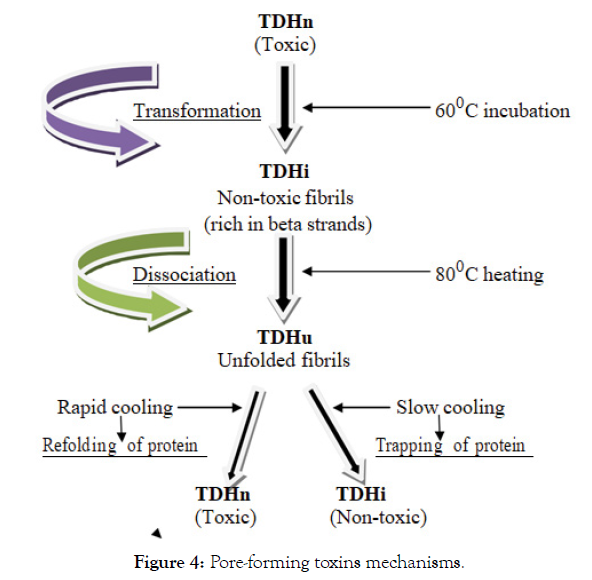
Figure 4: Pore-forming toxins mechanisms.
Haemolysins lyse erythrocytes, which results in the release of iron, an important growth factor for microorganisms, especially in pathogenicity, and during infections [196,197], as it is certain that numerous pathogenic microorganisms grow in the host by using haemin or haemoglobin as a source of iron [198-201]. Several fungal haemolysins have also thus, been proposed as virulence factors [202-204]. In addition to cell adherence, cytotoxicity and cell invasiveness, haemolysis also has an additional clinical significance, in being regarded as a virulence factor [205,206]. Furthermore, microbial haemolysins promote opportunistic infections and other clinical conditions, and also presented as risk factors in hospitals patients [202,207-209]. The expression of a haemolytic protein, with capabilities to lyse erythrocytes, has also been suggested as providing survival strategy for fungi during opportunistic infections [210]. The haemolysin, which enabled the fungus to disrupt blood cells, contained negatively charged domains that could also be detected in infected patients [166,211-213].
Research studies have shown that another application of fungal haemolysins has been their use as biomarkers for personal exposure to fungi or species-specific identification of opportunistic fungal diseases [166,214,215-217]. There is therefore, considerable interest in the development of diagnostic assays for detecting haemolysins as biomarkers of allergic and disseminated fungal exposure [166]. In actual fact, fungal haemolysins have been useful as biomarkers for exposure to indoor fungi because they can be measured in bodily fluids and environmental samples [202].
Extrapolations of the haemolysis screening assays in the determination of biosurfactants
According to Mulligan et al. [96] and Walter et al. [87], the technique of using blood agar plate haemolysis assay to screen for biosurfactant production on soluble substrates was shown to be quick and reliable. Some authors also believed that haemolysis screening method can be used to limit the number of samples, when selecting biosurfactant-producing microorganisms. In some cases, further screening for biosurfactant-producing microorganisms is only carried out, after screening for positive haemolytic activity [19]. The clear zone of haemolysis around the microbial colony on blood agar plates has commonly been related to the ability of the microbes to produce surfactants, while the diameter of the clear zone usually considered as a qualitative indicator of biosurfactant production [96,218]. It has however, been reported that haemolysis assay is not a specific method for biosurfactant production, since not all biosurfactants have haemolytic activities, basically due to presence of compounds other than biosurfactants [102,113]. Such other compounds include, virulence factors, toxins, and other lytic enzymes that can lyse erythrocytes [219]. It was also reported that biosurfactants that are poorly diffusible may not lyse erythrocytes nor cause haemolysis [220,221]. Furthermore, in some studies, haemolysis assay was found to exclude many good biosurfactantproducers, while in some reports, microbial strains with positive haemolytic activity were found to be negative for biosurfactant production [113]. There were a number of reports as well, which confirmed that microorganisms that were positive as biosurfactantproducing with the use other selection criteria, were negative for biosurfactant production when screened for haemolytic activity [88,113,121].
The poor specificity of haemolysis screening assay had also been confirmed, in that, it can give a lot of false-negative and falsepositive results [113,117]. In addition, it was reported that the diffusion restriction of surfactant can inhibit the formation of clearing zones on blood agar plates. Likewise, over-incubation of the blood agar plates may cause microbial overgrowth, which can lead to accumulation of microbial waste-products that may lyse the blood on blood agar plates; thereby, giving false appearance of biosurfactants, which are actually not present (http://www.encyclopedia.com/science/encyclopedias-almanacs-transcriptsand-maps/blood-agar-hemolysis-and-hemolytic-reactions). Until now, haemolytic microbial strains were generally believed to be biosurfactant-producers. Whereas, from the microbiological, clinical, pathophysiological and public health points of interpretations, the degree by which erythrocytes are haemolysed on blood-based culture media, is basically used to distinguish haemolytic and non-haemolytic microorganisms. Moreover, visualising the physical appearance of haemolysis on cultured blood agar plates has been used as a tool to determine the aetiologic (disease-causing) microbial species of various microbial infections [222].
In more recent times, biosurfactants have been generally considered as biodegradable, non-toxic (or minimally toxic), and eco-friendly/environmental-friendly compounds that are released by microorganisms [40,54,55,61,66,223-225]. But, apparently, most of the biosurfactants proposed in literature are reportedly produced by pathogenic microbes involved in pathogenesis, while relative levels of acute toxicity have also been recorded among significant numbers of surfactant-producing bacteria and fungi. The pathogenicity associated with haemolysis is therefore, a cause for concern, considering it being a commonly adopted biosurfactant potential and /or biosurfactant screening criterion. Therefore, contrary to the reports that biosurfactants-producing microorganisms used in some studies were generally recognised as safe (GRAS) [20], most of the microorganisms referred to as GRAS may still harbour one or more pathogenic/toxigenic/virulence factor(s), which can make them opportunistic pathogens [226].
Many well-characterised biosurfactant producers have been confirmed as pathogenic microbial species [24,203,227]. Conversely, haemolysis and haemolysins are specifically indicative of pathogenic and/or virulent or/and toxigenic status, rather than biosurfactant-production. The haemolytic action of certain bacteria and fungi on blood agar is so striking that haemolysis has been classified as very significant in clinical diagnosis of microbial importance. Due to the pathogenicity of some biosurfactantsproducing microorganisms [24,203,227,228], they were therefore, more recently, considered not appropriate for scaled- up production [25]. The detection of virulence genes coding for haemolysis and the determination of the antimicrobial resistance, in addition to the factors that contribute to pathogenicity and toxicity can contribute to better understanding of the need for better selection criteria of biosurfactants-producing microorganisms, and applications of their products [228], in the petroleum industries.
As earlier reported, literature on the production and analytical detection of biosurfactants is overwhelming, with assertions of high yields, and with mostly over-exaggerated estimates, due to the use of flawed or inaccurate analytical techniques [229]. However, none of the previous documented assertions on haemolysis assay, in biosurfactant determinations or contrariwise, highlighted the vivid microbiological and safety implications of haemolysis assay in biosurfactants-producing microorganisms. Based on the tremendous afore-mentioned intriguing justifications, haemolysis assay more appropriately identifies microbial strains with haemolytic (pathogenic/toxigenic/virulent) potentials. It can then be extrapolated that haemolysis assay (i.e., lyses of erythrocytes) on blood agar plates are more of diagnostic or determinative tools for microbial pathogenicity, rather than biosurfactants productions, and can therefore, not be conclusively confirmatory of biosurfactant-production, nor considered an appropriate selection criterion for biosurfactants. Thus, the likely or real pathogenicity, and/or virulence and toxicity of biosurfactants-producing microbes need to be appropriately assessed by haemolysis assay, prior to their potential applications in various petroleum industries, more especially, as they may be multi-antimicrobial resistant haemolytic.
From the petroleum industries perspectives, polycyclic aromatic hydrocarbons (PAHs) and naphthenic acids (NAs) are well-known to be toxic contaminants of environmental concern [230]. It is therefore, of necessity to ensure that additional hazardous concerns associated with petroleum activities are not introduced into the environment. A variety of microbial taxa are able to synthesize biosurfactants but it is ideal to isolate biosurfactants-producing microorganisms from appropriate and safe sources. From the ideal petroleum microbiology, public health, and hydrocarbon-processing points of view, the microbial strain profile and the ecological niche matter, as they determine the physiological status and metabolites production of the putative microorganisms. Therefore, it is proper to isolate biosurfactants-producing microorganisms from same or closely related ecological nich(es), for same physiological characteristics, extended survival, and maximal production of biosurfactant metabolites. Furthermore, toxic agents can cross microbial membranes [231], into the hosts; so, haemolysis assay, is hereby, suggested as, a highly determinative and qualitative screening assay indicative of the biosafety potentials, for the determination of pathogenic, toxigenic and/or virulent biosurfactant-producing microorganisms in the petroleum industries.
Conclusion
Biosurfactants are highly important microbial compounds of tremendous benefits but their significant public health concerns, especially regarding their haemolytic potentials serving as biosurfactant property are presently misconstrued. Bacterial and fungal haemolysins have been used as diagnostic tools, and/or biomarkers but microbial toxicity is undesirable in selected microbial candidates for various beneficial activities, such as, biosurfactantsproductions. Based on the intriguing afore-listed justifiable reasons, it can be noted and extrapolated that haemolysis assay; using blood agar is not so reliable, sensitive or suitable for determination of biosurfactant production, as it does not correlate particularly with specific characteristics of biosurfactants’ production. However, haemolysis assay is quite appropriately as, a reliable and sensitive safety bioassay, in routine monitoring, for pathogenic/toxigenic, and virulence determinations and regulations, as well as for selecting safe and environmental-friendly biosurfactant-producing microbial candidates.
Acknowledgements
This work was supported by the Klinkinberg Engineering Solutions Nigeria Limited [Grant number KESNL 0001, 2019].
REFERENCES
- Van Hamme JD, Singh A, Ward OP. Physiological aspects: Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol Adv 2006; 24:604-620.
- Otzen DE. Biosurfactants and surfactants interacting with membranes and proteins: Same but different? Biochimica et Biophysica Acta (BBA). Biomembranes. 2017; 1859:639-649.
- Karlapudi AP, Venkateswarulu TC, Tammineedi J, Kanumuri L, Ravuru BK, Dirisala VR, et al. Role of biosurfactants in bioremediation of oil pollution-A review. Petroleum. 2018; 4:241-249.
- Rahman PKSM, Mayat A, Joseph GHH, Randhawa KS, Relph LE, Armstrong MC. The role of microalgae in wastewater treatment. Front Bioeng Biotechnol. 2018; 169-188.
- Ghasemi A, Moosavi-Nasab M, Setoodeh P, Mesbahi G, Yousefi G. Biosurfactant production by lactic acid bacterium Pediococcus dextrinicus SHU1593 grown on different carbon sources: strain screening followed by product characterization. Scientific Reports. 2019.
- Jimoh AA, Lin J. Biosurfactant: A new frontier for greener technology and environmental sustainability. Ecotoxicol Environ Saf. . 2019; 184:109607.
- Park T, Jeon M-K, Yoon S, Lee KS, Kwon T-H. Modification of interfacial tension and wettability in oil–brine–quartz system by in situ bacterial biosurfactant production at reservoir conditions: implications for microbial enhanced oil recovery. Energy Fuels. 2019; 33: 4909-4920.
- Alfonso O, Teruel JA, Aranda FJ, Ortiz A. Effect of a dirhamnolipid biosurfactant on the structure and phase behaviour of dimyristoylphosphatidylserine model membranes. Colloids Surfaces B: Biointerfaces. 2020; 185: 110576.
- Singh AK, Sharma P. Disinfectant-like activity of lipopeptide biosurfactant produced by Bacillus tequilensis strain SDS21. Colloids and Surfaces B: Biointerfaces. 2020; 185:110514.
- Lenchi N, Kebbouche-Gana S, Servais P, Gana ML, Llirós M. Diesel biodegradation capacities and biosurfactant production in saline-alkaline conditions by Delftia sp NL1, isolated from an Algerian oilfield. Geomicrobiol J. 2020; 37(5).
- Cunha CD, do Rosario M, Rosado AS, Leite SGF. Serratia sp. SVGG 16: A promising bio-surfactant producer isolated from tropical soil during growth with ethanol-blended gasoline. Process Biochem. 2004; 39:2277-2282.
- Fenibo EO, Ijoma GN, Selvarajan R, Chikere CB. Microbial surfactants: the next generation multifunctional biomolecules for applications in the petroleum industry and its associated environmental remediation. Microorg. 2019; 7:581.
- Marcelino PRF, Gonçalves F, Jimenez IM, Carneiro BC, Santos BB, da Silva SS. Sustainable production of biosurfactants and their applications. Lignocellulosic Biorefining Technol. 2020; 159-183.
- Adeyemi BJ, Ajilo VI, Falode OA, Falobi EO. Enhancing oil recovery through high level data mining technology. Paper presented at the North African Technical Conference and Exhibition of The Society of Petroleum Engineers, Cairo, Egypt. 2013; 15-17.
- Ariech M, Guechi A. Assessment of four different methods for selecting biosurfactant producing extremely halophilic bacteria. Afr. J. Biotechnol. 2015; 14:1764-1772.
- De Almeida DG, Soares Da Silva RCF, Luna JM, Rufino RD, Santos VA, Banat IM, et al. Biosurfactants: promising molecules for petroleum biotechnology advances. Front Microbiol. 2016; 7:1718.
- Falode OA, Adeleke MA, Ogunshe AAO. Evaluation of indigenous biosurfactant-producing bacteria for de-emulsification of crude oil emulsions. Microbiol Res J Intern. 2017; 18: 1-9.
- Nguyen MT, Gotz F. Lipoproteins of Gram-Positive bacteria: key players in the immune response and virulence. Microbiol Mol Biol Rev. 2016; 80: 891-903.
- Santos DK, Rufino RD, Luna JM, Santos VA, Sarubbo LA. Biosurfactants: multifunctional biomolecules of the 21st century. Int J Mol Sci. 2016; 17:401.
- Gómez-Graña S, Perez-Ameneiro M, Vecino X, Pastoriza-Santos I, Perez-Juste J, Cruz JM, et al. Biogenic synthesis of metal nanoparticles using a biosurfactant extracted from corn and their antimicrobial properties. Nanomaterials. 2017; 7:139.
- Park T, Joo HW, Kim GY, Kim S, Yoon S, Kwon TH. Biosurfactant as an enhancer of geologic carbon storage: microbial modification of interfacial tension and contact angle in carbon dioxide/water/quartz systems. Front Microbiol. 2017; 8:1285.
- Datta P, Tiwari P, Pandey LM. Isolation and characterization of biosurfactant producing and oil degrading Bacillus subtilis MG495086 from formation water of Assam oil reservoir and its suitability for enhanced oil recovery. Bioresour Technol. 2018; 270: 439-448.
- Mouafo TH, Mbawala A, Ndjouenkeu R. Effect of different carbon sources on biosurfactants’ production by three strains of Lactobacillus spp. BioMed Research Int. 2018;15.
- Araújo HWC, Andrade RFS, Montero-Rodríguez D, Rubio-Ribeaux D, Alves da Silva CA, Campos-Takaki GM. Sustainable biosurfactant produced by Serratia marcescens 22 UCP 1549 and its suitability for agricultural and marine bioremediation applications. Microbial Cell Factories. 2019; 18:2.
- Tripathi L, Irorere VU, Marchant R, Banat IM. Marine derived biosurfactants: a vast potential future resource. Biotechnol Lett. 2018; 40:1441-1457.
- Kabir K, Deeni YY, Hapca SM, Moore L, Spiers AJ. Uncovering behavioural diversity amongst high-strength Pseudomonas spp. surfactants at the limit of liquid surface tension reduction. FEMS Microbiology Lett. 2019; 365: fny008.
- Decesaro A, Machado TS, Cappellaro ÂC, Rempel A, Margarites AC, Reinehr CO, et al. Biosurfactants production using permeate from whey ultrafiltration and bioproduct recovery by membrane separation process. J Surfactants Detergents. 2020; 23:539-551
- Karbalaei HH, Taghavi L, Hasani ZP. Induction of biosurfactant production from a native isolated moderately halophilic bacterium, Halomonas sp. MM93 in the presence of olive oil and study of its stability. JMBS. 2020; 11:21-28.
- Nikolova C, Gutierrez T. Use of microorganisms in the recovery of oil from recalcitrant oil reservoirs: current state of knowledge, technological advances and future perspectives. Front Microbiol. 2020; 10:2996.
- Marchant R, Banat IM. Microbial biosurfactants: Challenges and opportunities for future exploitation. Trends Biotechnol. 2012a; 30:558-565.
- Ławniczak L, Mareck R, Chrzanowski L. Contributions of biosurfactants to natural or induced bioremediation. Appl Microbiol Biotechnol. 2013; 97:2327-2339.
- Banat IM, Makkar RS, Cameotra SS. Potential commercial applications of microbial surfactants. Applied Microbiol Biotechnol. 2020; 53:495-508.
- Marchant R, Banat IM Biosurfactants: a sustainable replacement for chemical surfactants? Biotechnol Lett. 2012b; 34: 1597-1605.
- Vandana P, Singh D. Review on biosurfactant production and its application. Int. J. Curr. Microbiol App Sci. 2018; 7:4228-4241.
- Zajic JE, Gerson DF. Microbial extraction of bitumen from Athabasca oil sand, In Oil Sands and Oil Shale, (Ed.) Strausz O, New York, NY: USA. 1978:145-161.
- Falode OA, Aduroja OC. Development of local demulsifier for water-in-oil emulsion treatment. Int J Sci: Basic Appl Res. 2015; 24:301-320.
- Banat IM, Samarah M, Murad M, Horne R, Banerjee S. Biosurfactant production and use in oil tank clean up. World J Microbiol Biotechnol. 1991; 7:80-88.
- Kosaric N. Biosurfactants and their application for soil bioremediation. Food Technol Biotechnol. 2001; 39: 295-304.
- Jinfeng L, Lijun M, Bozhong M, Rulin L, Fangtian N, Jiaxi Z. The field pilot of microbial enhanced oil recovery in a high temperature petroleum reservoir. J Petrol Sci Eng. 2005; 48:265-271.
- Mulligan CN. Environmental applications for biosurfactants. Environ Pollut. 2005; 133:183-198.
- Omole O, Falode OA. Effects of mixing conditions, oil type and aqueous phase composition on some crude oil emulsions. J Appl Sci. 2005; 5: 873-877
- Shete AM, Wadhawa G, Banat IM, Chopade BA. Mapping of patents on bioemulsifier and biosurfactant: a review. J Sci Ind Res. 2006; 65: 91-115.
- Urum K, Grigson S, Pekdemir T, McMenamy S. A comparison of the efficiency of different surfactants for removal of crude oil from contaminated soils. Chemosphere 2006; 62:1403-1410.
- Franzetti A, Bestetti G, Caredda P, La Colla P, Tamburini E. Surface-active compounds and their role in the access to hydrocarbons in Gordonia strains. FEMS Microbiol Ecol. 2008; 63:238-248.
- Omole O, Falode OA. Stabilization of some crude oil emulsions by asphaltenes and reservoir fines. Int J Appl Chem. 2006a; 2: 29-43.
- Pornsunthorntawee O, Arttaweeporn N, Paisanjit S, Somboonthanate P, Abe M, Rujiravanit R, et al. Isolation and comparison of biosurfactants produced by Bacillus subtilis PT2 and Pseudomonas aeruginosa SP4 for microbial surfactant-enhanced oil recovery. Biochem Eng. 2008; 42:172-179.
- Das K, Mukherjee AK. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresource Technol. 2007; 98: 1339-1345.
- Haddadin MSY, Arqoub AAA, Reesh IA, Haddadin J. Kinetics of hydrocarbon extraction from oil shale using biosurfactant producing bacteria. Energ Convers Manage. 2009; 50:983–990.
- Lai CC, Huang YC, Wei YH, Chang JS. Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J Hazard Mater. 2009; 167:609-614.
- Owsianiak M, Chrzanowski Ł, Szulc A, Staniewski J, Olszanowski A, Olejnik-Schmidt AK, et al. Biodegradation of diesel/biodiesel blends by a consortium of hydrocarbon degraders: effect of the type of blend and the addition of biosurfactants. Bioresour Technol 2010; 100:1497-1500.
- Youssef N, Elshahed MS, McInerney MJ. Microbial processes in oil fields: culprits, problems, and opportunities, in Advances in Applied Microbiology, eds. Laskin A. I., Sariaslani S., Gadd G. M., Burlington: Academic Press, USA. 2009; 66: 141-251.
- Amaral PF, Coelho MA, Marrucho IM, Coutinho JA. Biosurfactants from yeasts: characteristics, production and application. Adv Exp Med Biol. 2010; 672:236-249.
- Assadi MM, Tabatabaee MS. Biosurfactants and their use in upgrading petroleum vacuum distillation residue: a review. Int J Env Res. 2010; 4:549-572.
- Banat IM, Franzetti A, Gandolfi I, Bestetti G, Martinotti MG, Fracchia L, et al. Microbial biosurfactants production, applications and future potential. Appl Microbiol Biotechnol. 2010; 87:427-444.
- Kiran GS, Thomas TA, Selvin J. Production of a new glycolipid biosurfactant from marine Nocardiopsis lucentensis MSA04 in solid-state cultivation. Colloids Surf. B: Biointerf. 2010; 78:8-16.
- Martha SVR, Ballinas CCPR, M deL, Torres-Muñoz JV, Rivera-Chavira BE, Nevárez-Moorillón GV. Selection of biosurfactant/bioemulsifier-producing bacteria from hydrocarbon-contaminated soil. Braz J Microbiol. 2010; 41:668-675.
- Perfumo A, Rancich I, Banat IM. Possibilities and challenges for biosurfactants use in petroleum industry, in Biosurfactants Advances in Experimental Medicine and Biology, ed. Sen R., New York, NY: Springer; USA. 2010. pp:135-145.
- Satpute SK, Banpurkar AG, Dhakephalkar PK, Banat IM, Chopade BA. Methods for investigating biosurfactants and bioemulsifiers. Crit Rev Biotechnol. 2010; 30:127-144.
- Falode OA, Ojumoola O. Evaluating the application of foam injection in enhancing oil recovery in unconsolidated sands. J Petrol Gas Engineering. 2015; 6:22-37.
- Falode OA, Adegoke SO. Production of biolubricants for oil well drilling applications. AU J Technol. 2015; 18:203-210.
- Sivapathasekaran C, Mukherjee S, Ray A, Gupta A, Sen R. Artificial neural network modeling and genetic algorithm based medium optimization for the improved production of marine biosurfactant. Bioresour Technol. 2010; 101:2884-2887.
- Viramontes-Ramos S, Cristina Portillo-Ruiz M, Ballinas-Casarrubias Mde L, Torres-Muñoz JV, Rivera-Chavira BE, Nevárez-Moorillón GV. Selection of biosurfactant/bioemulsifier-producing bacteria from hydrocarbon-contaminated soil. Braz J Microbiol. 2010; 41:668-675.
- Lima TMS, Fonseca AF, Leão BA, Mounteer AH, Tótola MR. Oil recovery from fuel oil storage tank sludge using biosurfactants. J Bioremed Biodegrad. 2011a; 2:12.
- Moldes AB, Paradelo R, Rubinos D, Devesa-Rey R, Cruz JM, Barral MT. Ex situ treatment of hydrocarbon-contaminated soil using biosurfactants from Lactobacillus pentosus. J Agric Food Chem. 2011; 59:9443-9447.
- Matsui T, Namihira T, Mitsuta T, Saeki H. Removal of oil tank bottom sludge by novel biosurfactant. JE1058BS. J Jpn Pet Inst. 2012; 55:138-141.
- Mohebali G, Kaytash A, Narges E. Efficient breaking of water/oil emulsions by a newly isolated de-emulsifying bacterium, Ochrobactrum anthropi strain RIPI5-1. Colloids Surf B. Biointerf. 2012; 98:120-128
- Moldes AB, Paradelo R, Vecino X, Cruz JM, Gudiña E, Rodrigues L, et al. Partial characterization of biosurfactant from Lactobacillus pentosus and comparison with sodium dodecyl sulphate for the bioremediation of hydrocarbon contaminated soil. BioMed Res Int. 2013:ID961842.
- Rocha e Silva NMP, Rufino RD, Luna JM, Santos VA, Sarubbo LA. Screening of Pseudomonas species for biosurfactant production using low-cost substrates. Biocatal Agric Biotechnol. 2013; 3:132-139.
- Sobrinho HBS, Luna JM, Rufino RD, Porto ALF, Sarubbo LA. Application of biosurfactant from Candida sphaerica UCP 0995 in removal of petroleum derivative from soil and sea water. Life Sci. 2013; 7:559–569.
- Mulligan CN, Sharma SK, Mudhoo A, Makhijani K. Green chemistry and biosurfactant research, in Biosurfactants: Research Trends and Applications, eds. Mulligan CN., Sharma SK., Mudhoo A. Boca Raton, FL: CRC Press, USA. 2014;1-30.
- Sarafzadeh P, Niazi A, Oboodi V, Ravanbakhsh M, Hezave AZ, Shahab Ayatollahi S Investigating the efficiency of MEOR processes using Enterobacter cloacae and Bacillus stearothermophilus SUCPM#14 (biosurfactant-producing strains) in carbonated reservoirs. J Pet Sci Eng. 2014; 113:46-53.
- Silva RCFS, Almeida DG, Rufino RD, Luna JM, Santos VA, Sarubbo LA. Applications of biosurfactants in the petroleum industry and the remediation of oil spills. Int J Mol Sci. 2014; 15:12523-12542.
- Vilela WF, Fonseca SG, Fantinatti-Garboggini F, Oliveira VM, Nitschke M. Production and properties of a surface-active lipopeptide produced by a new marine Brevibacterium luteolum strain. Appl Biochem Biotechnol. 2014; 174:2245-2256.
- Amani H, Kariminezhad H. Study on emulsification of crude oil in water using emulsan biosurfactant for pipeline transportation. Petrol Sci Technol. 2016; 34:216-222.
- Falode O, Oluwadero TA, Nwadike B, Fagade OE. Performance of biosurfactant produced from pineapple waste for improving oil recovery. Am Chem Sci J. 2016; 15:1-14.
- Kryachko Y. Novel approaches to microbial enhancement of oil recovery. J Biotechnol. 2018; 266:118-123.
- Lee DW, Lee H, Kwon BO, Khim JS, Yim UH, Kim BS, et al. Biosurfactant-assisted bioremediation of crude oil by indigenous bacteria isolated from Taean beach sediment. Environ Pollut. 2018a. 241:254-264.
- Lee DW, Lee H, Lee AH, Kwon BO, Khim JS, Yim UH, et al. Microbial community composition and PAHs removal potential of indigenous bacteria in oil contaminated sediment of Taean coast, Korea. Environ Pollut. 2018b; 234:503-512.
- Marchut-Mikolajczyk O, Drożdżyński P, Pietrzyk D, Antczak T. Biosurfactant production and hydrocarbon degradation activity of endophytic bacteria isolated from Chelidonium majus L. Microb Cell Fact. 2018; 17:171.
- Zhao F, Li P, Guo C, Shi RJ, Zhang Y. Bioaugmentation of oil reservoir indigenous Pseudomonas aeruginosa to enhance oil recovery through in-situ biosurfactant production without air injection. Bioresour Technol. 2018; 251:295-302.
- Marzorati S, Verotta L, Trasatti SP. Green corrosion inhibitors from natural sources and biomass wastes. Molecules. 2019; 24: 48.
- Patel S, Homaei A, Patil S, Daverey A. Microbial biosurfactants for oil spill remediation: pitfalls and potentials. Appl Microbiol Biotechnol. 2019; 103:27-37.
- Phetcharat T, Dawkrajai P, Chitov T, Mhuantong W, Champreda V, Bovonsombut S. Biosurfactant-producing capability and prediction of functional genes potentially beneficial to microbial enhanced oil recovery in indigenous bacterial communities of an onshore oil reservoir. Curr Microbiol. 2019; 76:382-391.
- Domingues PM, Oliveira V, Serafim LS, Gomes NCM, Cunha Â. Biosurfactant production in sub-oxic conditions detected in hydrocarbon-degrading isolates from marine and estuarine sediments. Int J Environ Res Public Health. 2020; 17: E1746.
- Płaza G, Achal V. Biosurfactants: eco-friendly and innovative biocides against biocorrosion. Int J Mol Sci. 2020; 21:2152.
- Sharma S, Pandey LM. Production of biosurfactant by Bacillus subtilis RSL-2 isolated from sludge and biosurfactant mediated degradation of oil. Bioresource Technol. 2020; 307:123261
- https://www.ncbi.nlm.nih.gov/books/NBK6189/
- Thavasi R, Sharma S, Jayalakshmi S. Evaluation of screening methods for the isolation of biosurfactant producing marine bacteria. J Phylogenetics Evol Biol. 2011; S1:001.
- Marchant R, Funston S, Uzoigwe C, Rahman PKSM, Banat IM. Production of biosurfactants from nonpathogenic bacteria, In Biosurfactants: Production and Utilization-Processes, Technologies, and Economics, Chap. 5 eds. Kosaric N., Sukan, Boca Raton: CRC Press. 2014; 73-82.
- Patowary K, Patowary R, Kalita MC, Deka S. Characterization of biosurfactant produced during degradation of hydrocarbons using crude oil as sole source of carbon. Front Microbiol. 2017; 8:279.
- Adnan M, Alshammari E, Ashraf SA, Patel K, Lad K, Patel M. Physiological and molecular characterization of biosurfactant producing endophytic fungi Xylaria regalis from the cones of Thuja plicata as a potent plant growth promoter with its potential application. Biomed Res Int. 2018; 7362148.
- Nayarisseri A, Singh P, Sing SK. Screening, isolation and characterization of biosurfactant producing Bacillus subtilis strain ANSKLAB03. Bioinformation. 2018; 14:304-314.
- Astuti DI, Purwasena IA, Putri RE, Amaniyah M, Sugai Y. Screening and characterization of biosurfactant produced by Pseudoxanthomonas sp. G3 and its applicability for enhanced oil recovery. J Petrol Exploration Production Technol. 2019; 9:2279-2289.
- Nierderhauser D, Bartell F. A corrected table for the calculation of boundary tensions by the pendent drop method. American Petroleum Institute, Baltimore, USA, 1950.
- Rosenberg E, Zuckerberg A, Rubinovitz C, Gutnick DL. Emulsifier of Arthrobacter RAG-I: isolation and emulsifying properties. Appl Environ Microbiol. 1979; 37:402-408
- Mulligan CN, Cooper D, Neufeld R. Selection of microbes producing biosurfactants in media without hydrocarbons. J Fermentation Technol. 1984; 62:311-314.
- Cooper D. Biosurfactants. Microbiol Sci. 1986; 3:145-149.
- Cooper D, Goldenberg B. Surface-active agents from 2 Bacillus species. Appl Environ Microbiol. 1987; 53:224-229.
- Peersson A, Molin G. Capacity for biosurfactant production of environmental Pseudomonas and Vibrionaceae growing on carbohydrates. Appl Microbiol Biotechnol. 1987; 26:439-442.
- Neu T, Poralla K. Emulsifying agents from bacteria isolated during screening for cells with hydrophobic surfaces. Appl Microbiol Biotechnol. 1990; 32:521-525.
- Jain DK, Collins-Thompson DL, Lee H, Trevors JT. A drop-collapsing test for screening surfactant-producing microorganisms. J Microbiol Methods. 1991; 13:271-279.
- Schulz D, Passeri A, Schmidt M, Lang S, Wagner F, Wray V, et al. Marine biosurfactants. 1. Screening for biosurfactants among crude-oil degrading marine microorganisms from the North-Sea. Z Naturforsch. 1991; 46:197-203.
- Siegmund I, Wagner F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol Tech. 1991; 5:265-268.
- Van der Vegt W, Van der Mei HC, Noordmans J, Busscher HJ. Assessment of bacterial biosurfactant production through axisymmetrical drop shape-analysis by profile. Appl Microbiol Biotechnol. 1991; 35:766-770.
- Morikawa M, Hirata Y, Imanaka T. A study on the structure function relationship of lipopeptide biosurfactants. Biochim Biophys Acta. 2000; 1488:211-218.
- Carrillo PG, Mardaraz C, Pitta-Alvarez SI, Giuliett AM. Isolation and selection of biosurfactant producing bacteria, World J Microbiol Biotechnol. 1996; 12:82-84.
- Pruthi V, Cameotra S. Rapid identification of biosurfact ant-producing bacterial strains using a cell surface hydrophobicity technique. Biotechnol Techniques. 1997; 11:671-674.
- Willumsen P, Karlson U. Screening of bacteria, isolated from PAH-contaminated soils, for production of biosurfactants and bioemulsifiers. Biodegrad. 1997; 7:415-423
- Bodour AA, Miller-Maier R. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J Microbiol Methods. 1998; 32:273-280.
- Huy NQ, Jin S, Amada K, Haruki M, Huu NB, Hang DT, et al. Characterization of petroleum-degrading bacteria from oil-contaminated sites in Vietnam. J Biosci Bioeng. 1999; 88:100-102.
- Tuleva BK, Ivanov GR, Christova NE. Biosurfactant production by a new Pseudomonas putida strain. Z Naturforsch [C]. J Biosci. 2002; 57:356-360.
- Bodour AA, Gerrero-Barajas C, Maier M. Structure and characterization of flavolipids, a novel class of biosurfactants produced by Flavolipid sp. strain MTN11. Appl Environ Microbiol. 2004; 10:1114-1120.
- Youssef NH, Duncan KE, Nagle DP, Savage KN, Knapp RM, McInerney MJ. Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004; 56:339-347.
- Bonilla M, Olivaro C, Caronal M, Vazquez A, Soubes M. Production and characterization of a new bioemulsifier from Pseudomonas putida ML2. J Appl Microbiol. 2005; 98:456-463
- Tadros T. Adsorption of surfactants at the air/liquid and liquid/liquid interfaces. In: Applied Surfactants: Principles and Applications. Weinheim: Wiley VCH. 2005; 81-82.
- Sarubbo LA, Moura de Luna J, de Campos-Takaki GM. Production and stability studies of the bioemulsifier obtained from a new strain of Candida glabrata UCP 1002. Electron J Biotechnol. 2006; 9: 4.
- Plaza G, Zjawiony I, Banat I. Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated bioremediated soils. J Petro Science Eng. 2006; 50:71-77.
- Chen C, Baker S, Darton R. The application of a high throughput analysis method for the screening of potential biosurfactants from natural sources. J Microbiol Meth. 2007; 70:503-510.
- Vaux D, Cottingham M. Method and apparatus for measuring surface configuration. Patent number: WO 2007/039729 A1, 2001.
- Maczek J, Junne S, Götz P. Examining biosurfactant producing bacteria—an example for an automated search for natural compounds. Application Note CyBio AG; 2007.
- Satpute SK, Bhawsar BD, Dhakephalkar PK, Chopade BA. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Indian J Marine Sci. 2008; 37:243-250.
- Belcher RW, Huynh KV, Hoang TV, Crowley DE. Isolation of biosurfactant-producing bacteria from the Rancho La Brea Tar Pits. World J Microbiol Biotechnol 2012; 28:3261-3267.
- Ogunshe AAO, Falode AO. Health and safety extrapolations of haemolysis assay in selecting biosurfactant-producing microorganisms in the petroleum industries and environment. International Conference on Hydrocarbon Science and Technology (ICHST) Theme: Oil and Beyond Oil: Strategies, Policies and Technologies to Assure National Energy Security. Petroleum Training Institute Conference Centre, Effurun, Delta State, Nigeria. 2019; 21-22.
- Mulligan CN, Mahmourides G, Gibbs BF. The influence of phosphate metabolism on biosurfactant production by Pseudomonas aeruginosa. J. Biotechnol 1989; 12:199-210.
- Robert M, Mercadé ME, Bosch MP, Parra JL, Espuny MJ, Manresa MA, et al. Effect of the carbon source on biosurfactant production by Pseudomonas aeruginosa 44T1. Biotechnol Lett. 1989; 11:871-874.
- Uchida Y, Misawa S, Nakahara T, Tabuchi T. Factors affecting the production of succinnoyl trehalose lipids by Rhodococcus erythropolis SD-74 grown on n-alkanes. Agric Biol Chem. 1989; 53:765-769.
- Davila AM, Marchal R, Vandecasteele JP. Sophorose lipid fermentation with differentiated substrate supply for growth and production phases. Appl Microbiol Biotechnol. . 1997; 47:496-501.
- Davis DA, Lynch HC, Varley J. The production of surfactin in batch culture by Bacillus subtilis ATCC 21332 is strongly influenced by the conditions of nitrogen metabolism. Enzyme Microb Technol. 1999; 25:322-329.
- Adamczak M, Bednarski W. Influence of medium composition and aeration on the synthesis of biosurfactants produced by Candida antarctica. Biotechnol Lett. 2000; 22:313-316.
- Sun W, Cao W, Jiang M, Saren G, Liu J, Cao J, et al. Isolation and characterization of biosurfactant-producing and diesel oil degrading Pseudomonas sp. CQ2 from Changqing oil field, China. RSC Advances. 2018; 8: 39710-39720.
- Bernheimer AW, Avigad LS. Nature and properties of a cytolytic agent produced by Bacillus subtilis. J General Microbiol. 1970; 61: 361-369.
- Morán AC, Martínez AM, Siñeriz F. Quantification of surfactin in culture supernatant by hemolytic activity. Biotechnol Lett. 2002; 24: 177-180.
- Afshar S, Lotfabad TB, Roostaazad R, Najafabadi AR, Noghabi AK. Comparative approach for detection of biosurfactant-producing bacteria isolated from Ahvaz petroleum excavation areas in south of Iran. Annalso of Microbiol. 2008; 58:555-560.
- Akintokun AK, Adebajo, SO, Akinremi, CA. Potential Biosurfactant-producing bacteria from pharmaceutical wastewater using simple screening methods in south-west, Nigeria. App Envi Res. 2017; 39: 41-54
- Smith ML, Gandolfi S, Coshall PM, Rahman PKSM. Biosurfactants: a Covid-19 perspective. Front Microbiol. 2020; 11:1341.
- Carter G. Isolation and identification of bacteria from clinical specimens. Diagnostic Procedures in Veterinary Bacteriology and Mycology. Charles C. Thomas (Ed.). 1984; 19-30.
- Banat IM. The isolation of a thermophilic biosurfactant producing Bacillus sp. Biotechnology Lett. 1993; 15: 591-594.
- Banat IM. Biosurfactant production and possible uses in microbial enhanced oil recovery and oil pollution remediation. Bioresource Technol. 1995a; 51: 1-12.
- Fiebig R, Schulze D, Chung JC, Lee ST. Biodegradation of polychlorinated biphenyls (PCBs) in the presence of a bioemulsifier produced on sunflower oil. Biodegrad. 1997; 8:67-75.
- Yonebayashi H, Yoshida S, Ono K, Enomoto H. Screening of microorganisms for microbial enhanced oil recovery process. Sekiyu Gakkaishi. 2000; 43:59-69.
- Tabatabaee A, Assadi MM, Noohi AA, Sajadian VA. Isolation of biosurfactant producing bacteria from oil reservoirs. Iranian J Env Health Sci Eng. 2005; 2: 6-12
- Dhail S. Isolation of potent biosurfactant producing bacteria from oil spilled marine water and marine sediments. Afr J Biotechnol. 2012; 11: 16751-16757.
- Ebrahimi A, Tashi N. Isolation of biosurfactant producing bacteria from poultry breast skin. Jundishapur J Nat Pharm Prod. 2012; 7: 93-96.
- Rodríguez-Rodríguez CE, Zúñiga-Chacón C, Barboza-Solano C. Evaluation of growth in diesel fuel and surfactants production ability by bacteria isolated from fuels in Costa Rica. Revista de la Sociedad Venezolana de Microbiología. 2012;32.
- Koim-Puchowska B, Kłosowski G, Mikulski D, Menka A. Evaluation of various methods of selection of b. subtilis strains capable of secreting surface-active compounds. PLoS One. 2019; 14:e0225108.
- Banat IM. Characterization of biosurfactants and their use in pollution removal – state of the art. Acta Biotechnologica. 1995b; 15:251-267.
- Lin SC. Biosurfactants: Recent advances. J Chem Tech Biotech. 1996; 66:109-120.
- https://www.asm.org/getattachment/0b25a2ee-06a6-41c1-bee4d086d3721b56/Blood-Agar-Plates-and-Hemolysis-Protocols.pdf
- https://microbiologyinfo.com/haemolysis-of-streptococci-and-its-types-with-examples/
- Gera K, McIver KS. Laboratory growth and maintenance of Streptococcus pyogenes (The Group A Streptococcus, GAS). Curr Protoc Microbiol. 2013; 30: 9D.2.1-9D.2.13.
- Favero D, Furlaneto-Maia L, França EJ, Góes HP, Furlaneto MC. Hemolytic factor production by clinical isolates of Candida species. Curr Microbiol. 2014; 68: 161-166.
- Mogrovejo DC, Perini L, Gostinčar C, Sepčić K, Turk M, Ambrožič-Avguštin J, et al. Prevalence of antimicrobial resistance and hemolytic phenotypes in culturable arctic bacteria. Front Microbiol. 2020; 11:570.
- Fujiwara A, Landau JW, Newcomer VD. Hemolytic activity of Rhizopus nigricans and Rhizopus arrhizus. Mycopathol Mycol Appl. 1970a; 40:131-138.
- Fujiwara A, Landau JW, Newcomer VD. Preliminary characterization of the hemolysin of Rhizopus nigricans. Mycopathol Mycol Appl. 1970b; 40:139-144.
- Macek P. Polypeptide cytolytic toxins from sea anemones (Actiniaria) FEMS Microbiol Immunol. 1992; 5:121-129.
- Sousa MV, Richardson M, Fontes W, Morhy L. Homology between the seed cytolysin enterolobin and bacterial aerolysins. J Prot Chem. 1994; 13:659-667.
- Anderluh G, Macek P. Cytolytic peptide and protein toxins from sea anemones (Anthozoa: Actiniaria) Toxicon. 2002; 40: 111-124.
- Berne S, Krizaj I, Pohleven F, Turk T, Macek P, Sepcić K. Pleurotus and Agrocybe hemolysins, new proteins hypothetically involved in fungal fruiting. Biochimica et Biophysica Acta. 2002; 1570: 153-159.
- Berne S, Sepcic K, Anderluh G, Turk T, Macek P, Poklar Ulrih N. Effect of pH on the pore forming activity and conformational stability of ostreolysin, a lipid raft-binding protein from the edible mushroom Pleurotus ostreatus. Biochemistry. 2005; 44: 11137-11147.
- Berne S, Lah L, Sepcic K. Aegerolysins: structure, function, and putative biological role. Protein Sci. 2009; 18:694-706.
- Schaufuss P, Steller U. Haemolytic activities of Trichophyton species. Med Mycol. 2003; 41:511-516.
- Ajay P, Brett J, Donald H. Fungal hemolysis. Medical Mycol. 2005; 51:1-16.
- Sher D, Fishman Y, Zhang M, Lebendiker M, Gaathon A, Mancheño JM, et al. Hydralysins, a new category of beta-pore-forming toxins in cnidaria. J Biol Chem. 2005; 280:22847–22855.
- Donohue M, Wei W, Wu J, Zawia NH, Hud N, De Jesus V, Schmechel D, et al. Characterization of nigerlysin, hemolysin produced by Aspergillus niger, and effect on mouse neuronal cells in vitro. Toxicol. 2006; 219: 150-155.
- Pipkin ME, Lieberman J. Delivering the kiss of death: progress on understanding how perforin works. Curr Opin Immunol. 2007; 19:301-308.
- Nayak AP, Green BJ, Beezhold DH. Fungal hemolysins. Med Mycol. 2013; 51:1-16.
- Ng TB, Cheung RCF, Wong JH, Chan YS, Dan X, Pan W, et al. Fungal proteinaceous compounds with multiple biological activities. Appl Microbiol Biotechnol. 2016; 100: 6601-6617.
- Furlaneto MC, Góes HP, Perini HF, Dos Santos RC, Furlaneto-Maia L. How much do we know about hemolytic capability of pathogenic Candida species? Folia Microbiol (Praha). 2018; 63:405-412.
- Gonzalez MR, Bischofberger M, Pernot L, van der Goot FG, Freche B. Bacterial pore-forming toxins: the (w)hole story? Cellular and Molecular Life Sci. 2008; 65:493-507.
- Aksimentiev A, Schulten K. Imaging α-hemolysin with molecular dynamics: ionic conductance, osmotic permeability and the electrostatic potential map. Biophysical J. 2005; 88:3745-3761. https://doi.org/10.1529/biophysj.104.058727
- http://www.ks.uiuc.edu/Research/hemolysin//
- Bi S, Wang W, Zhao Y, Ru S, Liu Y. Studies on hemolysis of hemolysin produced by Synechocystis sp. PCC 6803. J Ocean Univ China. 2011; 10:362-368.
- Ohnishi K, Nakahira K, Unzai S, Mayanagi K, Hashimoto H, Shiraki K, et al. Relationship between heat-induced fibrillogenicity and hemolytic activity of thermostable direct hemolysin and a related hemolysin of Vibrio parahaemolyticus. FEMS Microbiol Lett. 2011; 318:10-7.
- Pag U, Oedenkoven M, Papo N, Oren Z, Shai Y, Sahl HG. In vitro activity and mode of action of diastereomeric antimicrobial peptides against bacterial clinical isolates. J Antimicrob Chemoth. 2004; 53:230-239.
- Goebel W, Chakraborty T, Kreft J. Bacterial hemolysins as virulence factors. Antonie van Leeuwenhoek. 1988; 54:453-463.
- Parimon T, Li Z, Bolz DD, McIndoo ER, Bayer CR, Stevens DL, et al. Staphylococcus aureus α-hemolysin promotes platelet-neutrophil aggregate formation. J Infect Dis. 2013; 208:761-770.
- Ristow LC, Welch RA. Hemolysin of uropathogenic Escherichia coli: A cloak or a dagger? Biochimica et Biophysica Acta (BBA)-Biomembranes. 2016; 1858: 538-545.
- Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, et al. Staphylococcus aureus α-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis. 2012; 205: 807-817.
- Asam D, Mauerer S, Spellerberg B. Streptolysin S of Streptococcus anginosus exhibits broad-range hemolytic activity. Medical Microbiol Immunol. 2015; 204:227-237.
- Fan Y, Li Z, Li Z, Li X, Sun H, Li J, et al. Non-hemolysis of epidemic El Tor biotype strains of Vibrio cholerae is related to multiple functional deficiencies of hemolysin A. Gut Pathogens. 2019; 11(38).
- Arvizu MSM, Iturriaga MH, Escartín DF. Indicator and pathogenic bacteria in guacamole and their behavior in avocado pulp. J Food Safety. 2001; 21:233-244.
- Boguen R, Treulen F, Uribe P, Villegas JV. Ability of Escherichia coli to produce hemolysis leads to a greater pathogenic effect on human sperm. Fertil Steril. 2015; 103:1155-1161.
- Burnside K, Lembo A, de los Reyes M, Iliuk A, BinhTran NT, Connelly JE, et al. 2010. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS ONE. 2015; 5(6):e11071.
- Wiles TJ, Mulvey, MA. The RTX pore-forming toxin alpha-hemolysin of uropathogenic Escherichia coli: progress and perspectives. Future Microbiol. 2013; 8: 73-84.
- Perini L, Mogrovejo DC, Tomazin R, Gostinčar C, Brill FHH, Gunde-Cimerman N. Phenotypes associated with pathogenicity: their expression in arctic fungal isolates. Microorganisms. 2019; 7:E600.
- Richard KL, Kelley BR, Johnson JG. Heme uptake and utilization by Gram-negative bacterial pathogens. Front Cell Infect Microbiol. 2019; 9:81.
- Tee CB, Sei Y, Kajiwara S. Secreted hydrolytic and haemolytic activities of Malassezia clinical strains. Mycopathologia. 2019; 184:227-238.
- Wang C, Li Q, Lv J, Sun X, Cao Y, Yu K, et al. Alpha-hemolysin of uropathogenic Escherichia coli induces GM-CSF-mediated acute kidney injury. Mucosal Immunol. 2020; 13:22-33.
- Ebina K, Ichinowatari S, Yokota K. Studies on toxin of Aspergillus fumigatus. XXII Fashion of binding of Asp-hemolysin to human erythrocytes and Asp-hemolysin-binding proteins of erythrocyte membranes. Microbiol Immunol. 1985; 29:91-101.
- Yokota K, Ichinowatari S, Ebina K, Wakabayashi N. Studies on toxin of Aspergillus fumigatus XXI Site of binding of asp-hemolysin to erythrocytes and mechanism of inhibition of hemolysis. Jpn J Med Mycol. 1985; 26:70-73.
- Sepcic K, Berne S, Potrich C, Turk T, Macek P, Menestrina G. Interaction of ostreolysin, a cytolytic protein from the edible mushroom Pleurotus ostreatus, with lipid membranes and modulation by lysophospholipids. Eur J Biochem. 2003; 270:1199-1210.
- Parker MW, Feil SC. Pore-forming protein toxins: from structure to function. Prog Biophys Mol Biol. 2005; 88:91-142.
- Donohue M, Chung Y, Magnuson ML, Ward M, Selgrade MJ, Vesper S. Hemolysin chrysolysin from Penicillium chrysogenum promotes inflammatory response. Int J Hyg Environ Health. 2005; 208:279-285
- Dondapati SK, Wüstenhagen DA, Strauch E, Kubick S. Cell-free production of pore forming toxins: Functional analysis of thermostable direct hemolysin from Vibrio parahaemolyticus. Eng Life Sci. 2018; 18:140-148.
- Fukui T, Shiraki K, Hamada D, Hara K, Miyata T, Fujiwara S, et al. Thermostable direct hemolysin of Vibrio parahaemolyticus is a bacterial reversible amyloid toxin. Biochem. 2005; 44:9825-9832.
- Bullen JJ. The significance of iron in infection. Rev Infec Dis. 1981; 3:1127-1138.
- Kalidasan V, Joseph N, Kumar S, Hamat RA, Neela VK. Iron and virulence in Stenotrophomonas maltophilia: all we know so far. Front Cell Infect Microbiol. 2018; 8:401
- Manns JM, Mosser DM, Buckley HR. Production of a hemolytic factor by Candida albicans. Infect Immun. 1994; 62:5154-5156
- Watanabe T, Takano M, Murakami M, Tanaka H, Matsuhisa A, Nakao N, et al. Characterization of a haemolytic factor from Candida albicans. Microbiol. 1999; 145: 689-694.
- Luo G, Samaranayake LP, Yau JYY. Candida species exhibit differential in vitro hemolytic activities. Clin Microbiol. 2001; 39:2971-2974.
- Bairwa G, Jung WH, Kronstad JW. Iron acquisition in fungal pathogens of humans. Metallomics. 2017; 9: 215-227.
- Vesper SJ, Vesper MJ. Possible role of fungal hemolysins in sick building syndrome. Adv Appl Microbiol. 2004; 55:191-213.
- Rementeria A, Lopez-Molina N, Ludwig A, Vivanco AB, Bikandi J, Pontón J, et al. Genes and molecules involved in Aspergillus fumigatus virulence. Rev Iberoam Micol. 2004; 22:1-23.
- Majumdar T, Mullick JB, Bir R, Roy J, Sil SK. Determination of virulence factors and biofilm formation among isolates of vulvovaginal candidiasis. J Med Sci. 2016; 36:53-58.
- Payment P, Coffin E, Paquette G. Blood agar to detect virulence factors in tap water heterotrophic bacteria. Appl Environ Microbiol. 1994; 60:1179-1183.
- Kuroda H, Kuroda M, Cui L, Hiramatsu K. Subinhibitory concentrations of β-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol Lett. 2007; 268: 98-105.
- Rutqvist L. Studies on Aspergillus fumigatus; toxin production by different strains and serological comparison of the strains. Acta Vet Scand. 1965; 6:224-233.
- Ebina K, Ichinowatari S, Yokota K, Sakaguchi O. Studies on toxin of Aspergillus fumigatus XIX: Biochemical alterations of sera after Asp-hemolysin inoculation or Aspergillus infection in mice. Jpn J Med Mycol. 1984; 25:236-263.
- Vesper SJ, Vesper MJ. Stachylysin may be a cause of hemorrhaging in humans exposed to Stachybotrys chartarum. Infect Immun. 2002; 70:2065-2069.
- Aktas E, Yıgıt N. Hemolytic activity of dermatophytes species isolated from clinical specimens. J Mycol Med. 2015; 25:e25-30.
- Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance, FEMS Microbiol Revs. 2012; 36:288-305.
- Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Giannini MJM. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013; 62: 10–24.
- Figueiredo-Carvalho MHG, de Souza Ramos L, Barbedo LS, de Oliveira JCA, dos Santos ALS, Almeida-Paes R, et al. Relationship between the antifungal susceptibility profile and the production of virulence-related hydrolytic enzymes in Brazilian clinical strains of Candida glabrata. Mediators Inflamm. 2017; 8952878.
- CDC. Acute pulmonary hemorrhaging/hemosiderosis among infants: Cleveland, January 1993 - November 1994: Centers for Disease Control and Prevention, USA.
- Ebina K, Yokota K, Sakaguchi O. Studies on toxin of Aspergillus fumigatus XIV: relationship between Asp-hemolysin and experimental infection in mice. Jpn J Med Mycol. 1982; 23:246-252.
- Van Emon JM, Reed AW, Yike I, Vesper SJ. ELISA measurement of stachylysin in serum to quantify human exposures to the indoor mold Stachybotrys chartarum. J Occup Environ Med. 2003; 45:582-591.
- Wartenberg D, Lapp K, Jacobsen ID, Dahse HM, Kniemeyer O, Heinekamp T, et al. Secretome analysis of Aspergillus fumigatus reveals Asp-hemolysin as a major secreted protein. Int J Med Microbiol. 2011; 301: 602-611.
- Rodrigues L, Moldes A, Teixeira J, Oliveira R. Kinetic study of fermentative biosurfactant production by Lactobacillus strains. Biochem Eng J. 2006; 28:109-116.
- Balashova NV, Crosby JA, Al Ghofaily L, Kachlany SC. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infection and Immunity. 2006; 74: 2015-2021.
- Iqbal S, Khalid ZM, Malik KA. Enhanced biodegradation and emulsification of crude oil and hyperproduction of biosurfactants by a gamma ray induced mutant of Pseudomonas aeruginosa. Lett in Appl Microbiol. 1995; 21:176-179.
- Lin SC, Lin KG, Lo CC, Lin YM. Enhanced biosurfactant production by a Bacillus licheniformis mutant. Enzyme Microb Technol. 1998; 23:267-273.
- https://doi.org/10.1529/biophysj.104.058727
- Franzetti A, Tamburini E, Banat IM. Application of biological surface active compounds in remediation technologies. Adv Exp Med Biol. 2010; 672:121-134.
- Lima TMS, Procópio LC, Brandão FD, Leão BA, Tótola MR, Borges AC. Evaluation of bacterial surfactant toxicity towards petroleum degrading microorganisms. Bioresour Technol. 2011b; 102:2957-2964.
- Rincon-Fontan M, Rodrıguez-Lopez L, Vecino X, Cruz JM, Moldes AB. Adsorption of natural surface active compounds obtained from corn on human hair. RSC Adv. 2016; 6:63064-63070.
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000; 407:762-764.
- Flasz A, Rocha CA, Mosquera B. Differential pathogenicity of two bacterial strains: a clinical isolate and a biosurfactant-producing Pseudomonas aeruginosa ATCC 55925. Medical Pseudomonas aeruginosa Sci Res. 1997; 25: 461-464.
- Teker T, Albayrak G, Akayli T, Urku C. Detection of Haemolysin Genes as Genetic Determinants of Virulence in Lactococcus garvieae. Turk J Fish Aquat Sci. 2018; 19: 625-634.
- Marchant R, Banat IM. Protocols for measuring biosurfactant production in microbial cultures. Series: Springer Protocols Handbooks: Hydrocarbon and Lipid Microbiology Protocols, 2018.
- Reddy DO, Milliken CE, Foreman K, Fox J, Simpson W, Brigmon RL. Bioremediation of hexanoic acid and phenanthrene in oil sands tailings by the microbial consortium biotiger™. Bulletin of Environ Cont Toxicol. 2020; 104:253-258.
- Gill DM. Seven toxic peptides that cross cell membranes, In: J. Jeljaszewicz and T. Wadstrom (ed.), Bacterial toxins and cell membranes. Academic Press Inc., New York, USA. 1978; pp:291-332.
Citation: Adenike AOO, Falode OA (2021) The Intriguing Extrapolations of Haemolysis Assay as Screening Criterion for Selecting Biosurfactant- Producing Microorganisms in Petroleum Industries Process-Conditions. J Pet Environ Biotechnol. 8: 431.
Copyright: © 2021 Adenike AOO, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.