Indexed In
- Academic Journals Database
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 11, Issue 6
Synergistic Capability of Bacterial-fungal Co-culture to Degrade Drill Cutting Hydrocarbon
Desi Utami1, Donny Widianto1, Muhammad Saifur Rohman1, Sarmad Muhammad Soomar1, Julia Anggun1, Salman Ranani1 and Irfan Dwidya Prijambada*Received: 04-Jul-2019 Published: 12-Dec-2019
Abstract
Petroleum is a complex mixture of hydrocarbons. No single species of microorganisms is able to degrade all components of the petroleum. Mutually beneficial interaction of microorganisms in the form of a consortium is required during the process of oil degradation. One form of mutually beneficial interaction between fungi and bacteria is biofilm formed by bacteria on fungal surface. The formation of bacterial biofilm on fungal surface was reported to increase the synergistic action of the two microorganisms in destroying certain complex compounds. This work was aimed to assess the capacity of hydrocarbon degrading-bacterial biofilm on the surface of hydrocarbon degrading-fungal surface to degrade hydrocarbons derived from drill cuttings. The hydrocarbon degrading-soil bacteria and fungi were isolated from different area in Yogyakarta, Indonesia after enrichment. The ability of bacteria to form biofilm on the surface of fungal hyphae was examined under light microscope with 1000x magnification after the addition of lactophenol. The effect of microbial amendment in the form of biofilm, in comparison with the planktonic culture, on degradation of hydrocarbons derived from drill cuttings were assessed by measuring the extractable petroleum hydrocarbon. The results showed that co-culture between fungi and bacteria which one among them or both of them have low ability to degrade hydrocarbon may significantly improve their ability and the ability of the co-culture to degrade hydrocarbon has no relationship with the ability of the bacteria to form biofilm on the surface of the fungal hyphae.
Keywords
Bacteria; Biofilm; Fungi; Hydrocarbon
Introduction
Petroleum is a complex mixture of molecules, predominantly hydrocarbons. There are several hundred individual hydrocarbon chemicals that make up petroleum hydrocarbon. In high concentrations, these hydrocarbon molecules have a high toxicity against multiple organisms, including humans. Microorganisms play an important role in degrading petroleum compounds in polluted environments. The process of degradation of pollutant compounds by microorganisms is called biodegradation. Complexity petroleum hydrocarbon constituent causes the inability of a single species of microorganisms to degrade the overall components of the petroleum as each species of microorganisms can metabolize only a limited number of hydrocarbon substrates. Mutually beneficial interaction in the form of a consortium is required during the process of oil degradation [1-4].
Over the last few years the use of biofilms for biodegradation purposes has become a major focus of researchers. Biofilms are communities of microorganisms which grow and attach to a surface. The cells in the communities often bound one another by a matrix of extracellular polymeric substances (EPS). Biofilm formation is a way of microorganisms to protect themselves from physical and chemical environmental stress and to survive in nutrient-poor environments. Biofilms not only can be formed from the same types of microbes but they also can be formed from different microorganisms. One form of biofilm formed by different microbes are bacterial biofilms on fungal surface. In this type of biofilm, fungi act as a provider of solid surfaces for bacteria to stick to bacteria do not just use the surface of the fungal mycelium as a place of life but they also formed a special interaction with the fungus. The interaction basically provides an ideal environment for the formation of syntropic relationship between bacteria and fungi [5-9]. The formation of bacterial biofilm on fungal surface can provide protection for the bacteria from inhibitory effects of antimicrobial compounds, biocides, and a variety of other pressures, either chemical or physical. The polymer matrix in biofilms may also protect bacteria and fungi from the possibility of cells dehydration. Moreover, the formation of bacterial biofilm on fungal surface can also increase the synergistic action of the two microorganisms in destroying certain complex compounds. Isolated Pseudomonas species which formed biofilm was able to degrade crude oil much faster than its counterparts which could not form biofilm. Biofilm forming bacteria enable them to rapidly colonize the air-liquid interface by the production of EPS [10-12].
In this work, we aim to assess the capacity of hydrocarbon degradingbacterial biofilm on the surface of hydrocarbon degrading-fungal surface to degrade hydrocarbons derived from drill cuttings. The hydrocarbon degrading microorganisms, bacteria as well as fungi, were isolated from soils taken from different area in Yogyakarta, Indonesia after serial enrichment cultures. Isolates having ability to utilize crude oil as their carbon source were selected. The organisms were characterized and identified by morphological and biochemical test, as well as by sequencing of 16s rDNA, for bacteria, or Internal Transcribed Spacer (ITS) region1 dan 2, for fungi. The effects of microbial amendment in the form of biofilm, in comparison with the planktonic culture on degradation of hydrocarbons derived from drill cuttings were assessed.
Materials and Methods
Source of microorganisms, culture enrichment, and isolate characterization
Bacteria and fungi were isolated from various soils found in Yogyakarta, Indonesia. Soil samples (1 g) were dissolved into 9 ml of distilled water. The enrichment of drill cuttings hydrocarbondegrading microorganism was carried out by inoculating 10 ml Bushnell Haas minimal broth (per L of broth contains K2HPO4, 1 g; KH2PO4, 1 g; CaCl2, 0.2 g; MgSO4.7H2O, 0.02 g; and FeCl3, 0.005 g) containing 2% drill cuttings as its sole carbon source with 1 ml of the soil suspension. Drill cuttings were obtained from oil refinery unit of PT Pertamina (state-owned oil company) at Cilacap, Central Java, Indonesia. The enrichment process was carried out at room temperature with 120 rpm shaking. After seven days of incubation, the samples were diluted up to 10-3 and then were surface plated on PDA (Potato Dextrose Agar) for the isolation of fungi and NA (Nutrient Agar) for bacterial isolation. Colonies that appeared were then purified and its ability to degrade petroleum hydrocarbon was further examined quantitatively. The tests were conducted using Bushnell Haas minimal broth containing 2% petroleum hydrocarbon as the sole carbon source. The broth were inoculated with 1 loop of bacteria and incubated at room temperature with 120 rpm shaking for 7 days. After the incubation, residual hydrocarbon was extracted using benzene. The extractable residual petroleum hydrocarbons were quantified gravimetrically. Fungal and bacterial isolates having capability to degrade petroleum hydrocarbon were selected for further studies. Fungal isolates were characterized by their colonial and cellular morphology as well as by molecular identification. Colonial and cellular morphologies were observed by conventional dissecting microscope and compared the observation with standard keys, while molecular identification was conducted using primary part of the internal transcribed spacer (ITS). Identification of the selected bacteria was carried out based on colony morphology, biochemical characteristics following Bergey’s manual of determinative bacteriology and by 16s r-DNA genes sequencing [13-15]. Bacterial growth was measured by using spectrophotometer.
DNA extraction and PCR amplification
Fungi were cultivated in Potato Dextrose Broth (PDB) for 6-8 days at room temperature with agitation at 170 rpm. DNA extraction was performed using the method developed by Lee et al. with some modifications [16]. Fungal mycelium were collected using filter paper, and 1 g of the collected mycelium were ground using mortar after an addition of 1 ml lysis buffer (50 mM Tris-HCl, 50 mM EDTA, 3% SDS, 1% 2-mercaptoethanol). The lysate was then transfered into 1.5 ml microtubes and incubated in a water bath for 1 hour at 65°C. After centrifugation at 3400 rpm for 5 min at room temperature, the supernatant was transfered into a new microtubes and an equal volume of SEVAG (chloroform:isoamyl alcohol, 24:1, vol/vol) was then added. The mixture was then vortexed for a few secs, followed by centrifugation at 12000 rpm for 10 min at room temperature. Supernatant (580 μl) was taken and put into a new microtube, and sodium acetate 3 M (20 μl) was then added, followed by addition of 600 μl cold isopropanol. The mixture was then gently mixed by inverting the tube, and centrifuged at 12000 rpm for 2 min at room temperature. The supernatant was discarded and the tube containing pellet was stood in an inverted position for 1 min on a paper towel to allow all of the fluid to drain away. The pellet was then suspended in 300 μl elution buffer. RNase (100 mg/ml) was then added at 1 μl and the suspension was then incubated at 65°C for 15 min. After the incubation, 250 μl ammonium acetate (7.5 M) was added, mixed gently by inverting the tube, and then centrifuged at 12000 rpm for 5 min at room temperature. The supernatant was then transfered to a new microtube, and 750 ml isopropanol was then added. The mixture was then centrifuged at 12000 rpm for 2 min, and the supernatant was then discarded. Cold 100% ethanol (1 ml) was added to the pellet, vortexed for a few sec, and then centrifuged at 12000 rpm for 2 min at room temperature. The supernatant was then discarded, and the pellet was then rinsed with 1 ml 70% cold ethanol. The mixture was again centrifuged at 12000 rpm for 2 min at room temperature, and the supernatant was then discarded. The tube was inverted for 1 min to drain. The tube was further dried in an oven at 50°C for 15 min at most; the DNA pellet was then resuspended in 30-50 μl of TE, and then stored at -20°C.
The internal transcribed spacer region 1 (ITS1) was amplified by using GoTaq Green master mix (Promega). Specific ITS1 (5'-TCTGTAGGTGAACCTGCCG-3'), and ITS2 (5'TCCTCCGCTTATTGATATGC-3') primers pair were used for amplification [11]. Amplification was performed in a thermal cycler (GeneAmp PCR System 9700) using the following program: initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 52°C for 30s and extension at 72°C for 1 min. For the last cycle, the extension step was extended for another 5 min.
Bacterial isolates were cultivated in Nutrient Broth (NB) for 18-24 h at room temperature with agitation at 170 rpm. DNA extraction was performed using the method of Wilson with some modification [17]. Cells were harvested by centrifugation at 12000 rpm for 2 min at room temperature and the supernatant was discarded. The pellet was resuspended in 410 μl TE (Tris-EDTA) buffer by repeat pipetting, 50 μl lyzozim was then added and the mixture is incubated at 37°C for 30 min. The tube was inverted for gently mixing every 10 min. Addition of 30 μl of 10% sodium dodecyl sulfate and 3 μl of 20 mg/ml proteinase K followed by incubation at 37°C for 30 min with gently mixing every 10 min to ensure cells lysis. Sodium chloride 5 M (100 μL) and CTAB 10% (100 μl) were then added to the mixture, and the incubation was continued at 65°C for 15 min. An equal volume of SEVAG was then added, mixed thoroughly, and then centrifuged at 12000 rpm for 5 min at room temperature. Viscous supernatant was transfered to a new microtube. Cold isopropanol (0.6 volume) was then added, mixed thoroughly and then incubated overnight at 20°C. The mixture was then centrifuged 12000 rpm for 15 min at room temperature. The pellet was then rinse with 70% ethanol, incubated at 4°C for 1 h, and then centrifuged at 12000 rpm for 5 min at room temperature to repellet it. The supernatant was carefully removed, and the tube was inverted for 1 min to drain. The tube was further dried in an oven at 50°C for 15 min at most; the DNA pellet was then resuspended in 50 μl of TE buffer, and stored at 20°C.
For the amplification of 16s rDNA gene, F8 (5‘AGAGTTTGATCCTGGCTCAG-3’), and R1493 (5‘- ACGGCAACCTTGTTACGACC-3’) pair of primers were used with GoTaq Green master mix (Promega) kit. Amplification was performed in a thermal cycler (GeneAmp PCR System 9700) using the following program: initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 30 s and extension at 72°C for 1 min. For the last cycle, the extension step was extended for another 5 min.
The PCR-amplified DNAs were sent to 1st BASE, Singapore for sequencing. The sequences obtained were analyzed by BLAST online comparison (http:// www.ncbi.nlm.nih.gov) for identification based on homology comparison with NCBI databases [18,19].
Examination of the capability to form biofilm
The capability of bacterial isolates to form biofilm on the surface of fungal isolates was examined by co-culturing the bacterial and fungal isolates in Bushnell Haas minimal broth with glucose as its carbon source. Prior to the co-culturing process, the density of fungal spores and bacteria cells were calculated. Fungal spores (105) were inoculated into 50 ml Bushnell Haas minimal broth with pH 7.0 ± 0.2 and incubated at room temperature with 120 rpm shaking for 2 d until the fungi start to form small clumps. Bacterial cells (105) were then inoculated into the broth, and incubation at room temperature were continued with 80 rpm shaking. Biofilm formations were observed every day after the bacterial inoculation using light microscope with 1000x magnification after the addition of lactophenol. Biofilm formation was characterized by the attachment of bacterial cells on fungal hyphae.
Examination for the capability of bacterial biofilm on fungal surface to degrade petroleum hydrocarbon
Biofilm (0.2 g) was taken and sprayed with sterile distilled water to remove the free living bacterial cell, then inoculated into Bushnell Haas minimal broth containing 2% drill-cuttings and incubated at room temperature for 10 days with shaking at 120 rpm. Residual petroleum hydrocarbons in the broth were extracted using benzene and separated from the broth using separatory funnel. The extracted residual petroleum hydrocarbons were transferred into a petri dish and allowed to dry for 24 h. The extracted residual petroleum hydrocarbons deposited in the petri dish was then weighed. Petroleum hydrocarbon degradation ability was calculated from the difference between the weight of residual petroleum hydrocarbons extracted from the inoculated broth and uninoculated broth.
Examination for the capability of bacterial and fungal constituents of the biofilm on fungal surface to degrade petroleum hydrocarbon
To compare the ability of each bacterial and fungal constituent of the bacterial biofilm on fungal surface as well as the co-culture of bacteria-fungus that does not form biofilms on the degradation of petroleum hydrocarbons, the number of bacterial cell and fungal biomass in the biofilm should be determined. For that purpose, the number of bacteria on the biofilms which have been formed was counted by the pour plate method on Bushnell Haas minimal agar containing 2% glucose added with 200 ppm ketoconazole as an antifungal to inhibit the growth of fungi [20]. Our preliminary study showed that addition of 200 ppm ketoconazole to Bushnell Haas minimal agar containing 2% glucose did not significantly interfere the number colonies appeared on the plates from the same inocula. The fungal dry biomass was determined gravimetrically after 36 h drying at 80°C. The relationship between the number of bacteria cultured in a growth medium with their optical density was then determined. Inoculum of bacterial constituent of the biofilm containing an equal number of bacteria with that of the biofilm was inoculated into Bushnell Haas minimal broth containing 2% drill-cuttings and incubated at room temperature for 10 days with shaking at 120 rpm. On the other hand, inoculum of fungal constituent of the biofilm containing an equal amount of fungal biomass with that of the biofilm was inoculated into the same medium and incubated in the same way. After the incubation period, the ability of oil degradation was determined.
Statistical analysis
The data are the means of three replicates. Two factor analysis of variance (ANOVA) were performed using Microsoft Excel software to determine the level of significance at p<0.05. The differences between groups were analysed by Duncan’s Multiple Range (DMR) Test [21].
Results and Discussion
Isolation and selection of drill cutting hydrocarbon degrading bacteria and fungus Bacteria and fungi were isolated from various sources such as compost, peat, and soils found in Yogyakarta, Indonesia. Each source has different lignin content. Microorganisms were isolated after enrichment in minimal medium containing petroleum as the sole carbon source. Seven bacterial and three fungal isolates that were able to degrade petroleum hydrocarbon were obtained.
Bacterial isolates were then characterized morphologically, physiologically as well as genetically. The seven bacterial isolates were identified as Ochrobactrum anthropi, Pseudomonas aeruginosa, P. geniculata, Alcaligenes faecalis, Bacillus subtilis, B. cereus, and Acinetobacter baumannii. The fungal isolates were characterized by their colonial and cellular morphology as well as by molecular identification. The three fungal isolates were identified as Aspergillus niger, Eupenicillium javanicum and Penicillium sp. Capability of the bacterial and fungal isolates to degrade hydrocarbons contained in drill cuttings were shown in Table 1.
Capability of the bacterial isolates to form biofilm on the surface of fungal isolates
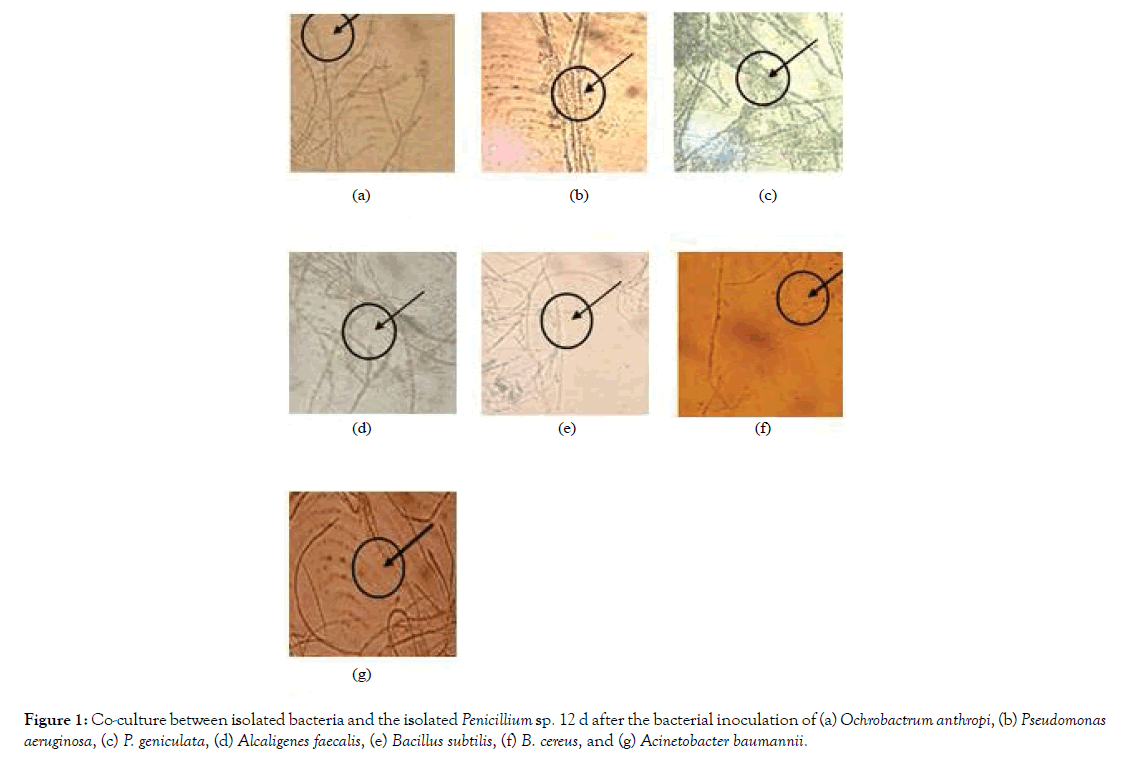
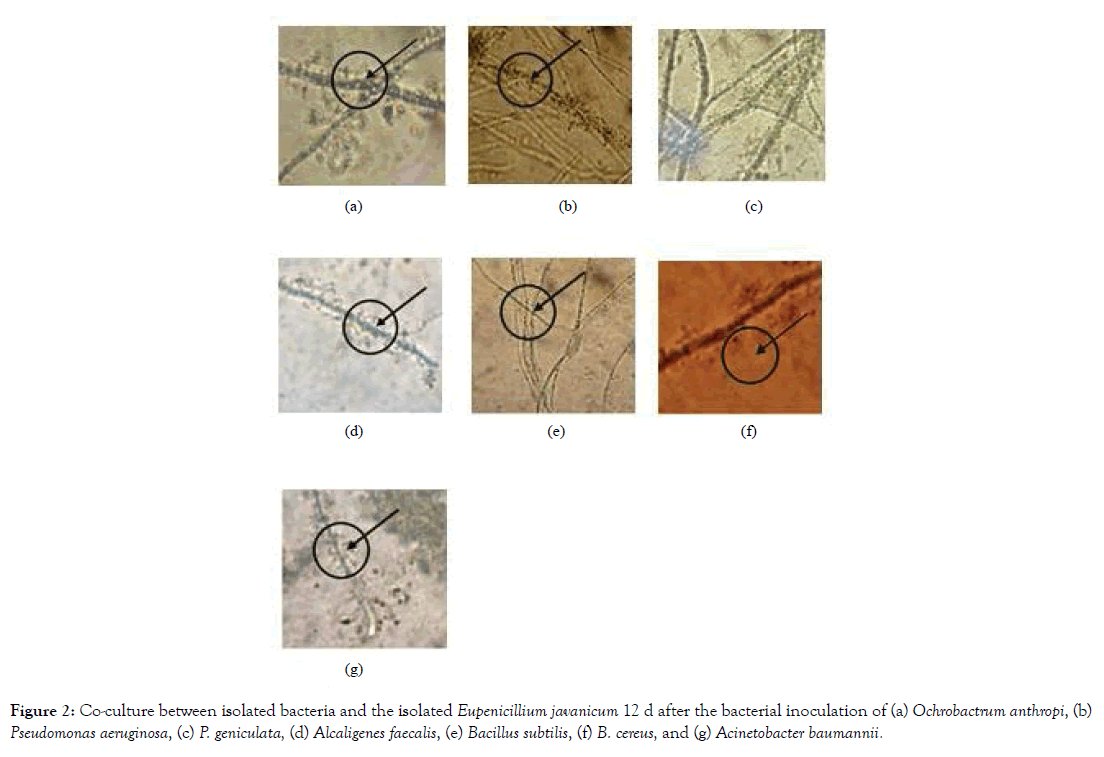
The obtained bacterial isolates were then examined for their capability to form biofilm on the surface of the obtained fungal isolates. After 7 days from inoculation and incubation bacterial isolate begins stick on surface fungus mycelium. No bacterial isolates were observed to be able to be co-cultured with Aspergillus niger. Co-culture on the surface of the isolated Penicillium sp. 12 d after the bacterial inoculations were shown in Figure 1, while the ones on Eupenicillium javanicum were shown in Figure 2.
Figure 1: Co-culture between isolated bacteria and the isolated Penicillium sp. 12 d after the bacterial inoculation of (a) Ochrobactrum anthropi, (b) Pseudomonas aeruginosa, (c) P. geniculata, (d) Alcaligenes faecalis, (e) Bacillus subtilis, (f) B. cereus, and (g) Acinetobacter baumannii.
Figure 2: Co-culture between isolated bacteria and the isolated Eupenicillium javanicum 12 d after the bacterial inoculation of (a) Ochrobactrum anthropi, (b) Pseudomonas aeruginosa, (c) P. geniculata, (d) Alcaligenes faecalis, (e) Bacillus subtilis, (f) B. cereus, and (g) Acinetobacter baumannii.
As shown in Figure 1, although all of the bacterial isolates can be co-cultured with the isolated Penicillium sp., only Pseudomonas aeruginosa and P. geniculata were able to form biofilms on the on the surface of the isolated Penicillium sp. The rest of the bacterial isolates can be co-cultured with Penicillium sp. without forming biofilm of the surface of the fungal mycellia. On the other hand, all of the seven bacterial isolates were able to form biofilms on the surface of the isolated Eupenicillium javanicum (Figure 2).
Examination for the capability of fungal-bacterial association to degrade petroleum hydrocarbon
The ability of the co-cultured between Penicillium sp. as well as Eupenicillium javanicum with seven bacterial isolates to degrade petroleum hydrocarbon was then examined. The results of the examination were shown in Table 2. Table 2 shows that inoculation of the petroleum containing Bushnell Haas minimal broth with only Penicillium sp. inoculum produced significant reduction on the amount of extractable petroleum hydrocarbon compared to the uninoculated one (control). It indicates the ability of Penicillium sp. to degrade petroleum hydrocarbon. However, inoculation with only Eupenicillium javanicum inoculum has no significant effect. The result is in accordance with the result of the selection process shown in Table 1, i.e. Eupenicillium javanicum has much lower ability to degrade petroleum hydrocarbon compared to that of Penicillium sp. In the absence of fungal inoculum, only petroleum containing broth inoculated with Pseudomonas aeruginosa, Alcaligenes faecalis, and Acinetobacter baumanii produced significant reductions on the amount of extractable petroleum hydrocarbon, which mean that those three bacteria have the ability to degrade petroleum hydrocarbon. However, in the presence of fungal inocula, both Penicillium sp. as well as Eupenicillium javanicum, inoculation of petroleum containing broth with Ochrobactrum anthropi, Pseudomonas geniculata, and Bacillus subtilis also resulted in a significant decrease on the amount of extractable petroleum hydrocarbon. It indicates that O. anthropi, P. geniculata, and B. subtilis are able to work synergeticaly with the fungi to degrade petroleum hydrocarbon. Only inoculation with Bacillus cereus, neither in the presence nor in the absence of fungal inuculum, has no effect on the amount of extractable petroleum hydrocarbon.
| Isolates | Benzene Extractable- Residual Hydrocarbon (mg) | ||
|---|---|---|---|
| No isolate (control) | 155.25 | ± | 0.35 |
| Ochrobactrum anthropi | 92.63 | ± | 7.53 |
| Pseudomonas aeruginosa | 57.97 | ± | 9.83 |
| Pseudomonas geniculata | 81.5 | ± | 12.19 |
| Alcaligenes faecalis | 95.67 | ± | 13.36 |
| Bacillus subtilis | 70.87 | ± | 8.42 |
| Bacillus cereus | 97.93 | ± | 5.63 |
| Acinetobacter baumanii | 59.97 | ± | 5.4 |
| Aspergillus niger | 58.4 | ± | 0.41 |
| Eupenicillium javanicum | 118.1 | ± | 1.4 |
| Penicillium sp. | 31.2 | ± | 0.52 |
Table 1: Benzene extractable-residual hydrocarbon in Bushnell Haas minimal broth containing 2% petroleum hydrocarbon inoculated with isolates.
| Treatments | Extractable petroleum hydrocarbon (g) from Minimal Medium inoculated with | |||||||
|---|---|---|---|---|---|---|---|---|
| O. anthropi | P. aeruginosa | P. geniculata | A. faecalis | B. subtilis | B. cereus | A. baumannii | No bacteria | |
| With Penicillium sp. inoculant | 0,694efg | 0,739defg | 0,758cdef | 0,762cde | 0,779cde | 0,887ab | 0,776cde | 0,811bcd |
| With Eupenicillium javanicum inoculant | 0,706efg | 0,662fg | 0,650g | 0,658g | 0,682efg | 0,834abcd | 0,650g | 0,826abcd |
| No fungal inoculant | 0,892ab | 0,769cde | 0,846abc | 0,797bcde | 0,860abc | 0,856abc | 0,768cde | 0,932a |
| Note: Different letters imply that there is a significant difference at p=0.05 *Shaded back groud indicates the ability of bacterial isolates to form biofilm on the surface of fungal mycelia |
||||||||
Table 2: Extractable petroleum hydrocarbon from Bushnell Haas minimal broth inoculated with Penicillium sp. or Eupenicillium javanicum-bacterial association and their bacterial as well as fungal constituents to degrade petroleum hydrocarbon.
If compared with the ability of Penicillium sp., P. aeruginosa, A. faecalis, and A. baumanii when inoculated alone, co-inoculations of the petroleum containing Bushnell Haas minimal broth with Penicillium sp. and P. aeruginosa, A. faecalis, or A. baumanii did not show any significant improvement on the ability of both fungal and bacterial inoculant to degrade petroleum hydrocarbon. However, co-inoculation of the petroleum containing Bushnell Haas minimal broth with Penicillium sp. and Ochrobactrum anthropi, Pseudomonas geniculata, or Bacillus subtilis resulted in significant improvement of the ability to degrade petroleum hydrocarbon compared to the ones inoculated only with bacterial inocula. The above results indicate that co-inoculation with Penicillium sp. could not further improve the ability of bacteria which have ability to degrade hydrocarbon. On the contrary, for bacteria which have low22 ability to degrade hydrocarbon, the co-inoculation with Penicillium sp. may significantly improve their ability. Penicillium sp. has been reported able to degrade the hydrocarbon because it uses crude oil as a source of carbon and energy and metabolized to CO2 and biomass [22]. Fungal hyphae are not solely as film to help the attachment but perform numerous actions, such as providing nutrients for bacteria [23] and facilitate bacteria to move with hyphae to access the nutrients [24].
Table 2 also shows that co-inoculation of the petroleum containing Bushnell Haas minimal broth with Eupenicillium javanicum, that has low ability to degrade hydrocarbon, and all bacterial inocula, except Bacillus cereus, significantly reduce the amount of extractable petroleum hydrocarbon compared to the ones inoculated only with bacterial inocula. It indicates that Eupenicillium javanicum could work synergistically with bacterial inocula in degrading the hydrocarbon. It was reported, that filamentous fungi are usually able to attach to the hydrophobic surface through the construction of hydrophobic proteins [25]. Formation of the Eupenicillium javanicum with 7 bacteria and its architecture helped in the effective transport of the bacterium following degradation of its surface by enzymatic reactions. Other research stated that various chemical catalyst produced by fungi attract the bacteria by chemotactic response such as expression of distinct genes consequent to association with the fungal cells [26], while Bacillus subtilis has been observed to shift through soil toward fungal spores as a chemotactic response [27] and Pseudomonas fluorescens has been found to induce its trehalose usage genes when bared to fungal culture supernatant [28].
Comparing Tables 3 and 4, it showed that co-cultivation between Eupenicillium javanicum and Ochrobactrum anthropi, Pseudomonas geniculata, Bacillus subtilis, and Alcaligenes faecalis increased their abilities exceeding the sum of their each ability to reduce hydrocarbon content in the Bushnell Haas medium. The same case can also be seen in the co-cultivation between Penicillium sp. and Ochrobactrum anthropi.
| Treatments | O. anthropi | P. aeruginosa | P. geniculata | A. faecalis | B. subtilis | B. cereus | A. baumannii | No bacteria |
|---|---|---|---|---|---|---|---|---|
| With Penicillium sp. inoculant | 0,226 | 0,270 | 0,282 | 0,274 | 0,249 | 0,098 | 0,282 | 0,106 |
| With Eupenicillium javanicum inoculant | 0,237 | 0,192 | 0,174 | 0,170 | 0,153 | 0,045 | 0,155 | 0,121 |
| No fungal inoculant | 0,112 | 0,213 | 0,145 | 0,144 | 0,044 | 0,076 | 0,216 | - |
Table 3: Differences between hydrocarbons extracted from Bushnell Haas minimal broth inoculated with Penicillium sp. or Eupenicillium javanicum-bacterial association and their bacterial as well as fungal constituents with control (without bacterial or fungal inoculation).
| Treatments | O. anthropi | P. aeruginosa | P. geniculata | A. faecalis | B. subtilis | B. cereus | A. baumannii |
|---|---|---|---|---|---|---|---|
| With Penicillium sp. inoculant | 0,218 | 0,319 | 0,251 | 0,250 | 0,150 | 0,182 | 0,322 |
| With Eupenicillium javanicum inoculant | 0,233 | 0,334 | 0,266 | 0,265 | 0,165 | 0,197 | 0,337 |
Table 4: Addition of the ability to reduce hydrocarbons by single inoculation of bacteria and fungi.
The case of co-inoculation with Ochrobactrum anthropi and Penicillium sp., in which the bacterial isolate is unable to form biofilm on the surface of fungal mycelium, and co-inoculation with Bacillus cereus and Eupenicillium javanicum in which the bacterial isolate is able to form biofilm on the surface of fungal mycelium, prevent us to conclude that the ability of bacterial isolates to form biofilm on the surface of fungal biomass may has an effect on their ability to degrade hydrocarbon.
Conclusion
Co-culture between fungi and bacteria which one among them or both of them have low ability to degrade hydrocarbon may significantly improve their ability and the ability of the co-culture to degrade hydrocarbon has no relationship with the ability of the bacteria to form biofilm on the surface of the fungal hyphae.
REFERENCES
- Matar S, Hatch LF. Chemistry of petrochemical processes. 2nd edn. Gulf Publishing Company, Texas. 2000.
- Alexander M. Biodegradation and bioremediation. Academic Press, San Diego. 1994.
- Leahy JG, Colwell RR. Microbial degradation of hydrocarbons in environment. Microbiol Rev. 1990;54:305-315.
- Olajire AA, Essien JP. Aerobic degradation of petroleum components by microbial consortia. J Pet Environ Biotechnol. 2014;5:195-217.
- Heydorn A, Ersbøll BK, Hentzer M, Parsek MR, Givskov M, Molin S. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146:2409–2415.
- O'toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49-79.
- Hogan DA, Wargo MJ, Beck N. Bacterial biofilms on fungal surfaces. In: Kjelleberg S, Givskov M. (eds.) The biofilm mode of life: Mechanisms and adaptations. Horizon Bioscience, Norfolk, U.K. 2007.
- Artursson V, Jansson JK. Use of bromodeoxyuridine immunocapture to identify active bacteria associated with arbuscular mycorrhizal hyphae. Appl Environ Microbiol. 2003;69:6208–6215.
- Davey ME, O'toole GA. Microbial biofilms: From ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867.
- Hogan DA, Kolter R. Pseudomonas-candida interactions: An ecological role for virulence factors. Science. 2002;296:2229–2232.
- Dasgupta D, Ghosh R, Sengupta TK. Biofilm-mediated enhanced crude oil degradation by newly isolated Pseudomonas species. ISRN Biotechnol. 2013;2013:250749.
- Meliani A, Bensoltane A. Enhancement of hydrocarbons degradation by use of Pseudomonas biosurfactants and biofilms. J Pet Environ Biotechnol. 2014;5:168-174.
- Barnett HL, Hunter BB. Illustrated Genera of Imperfect Fungi. 4th edn. APS Press. St Paul, Minnesota. 1998.
- Kim JY, Yeo SH, Baek SY, Choi HS. Molecular and morphological identification of fungal species isolated from Bealmijang Meju. J Microbiol Biotechnol. 2011;21:1270-1279.
- Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761-2764.
- Lee SB, Milgroom MG, Taylor JW. A rapid, high yield mini-prep method for isolation of total genomic DNA from fungi. Fungal Genetics Reports. 1988;35:23–24.
- Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, (eds). Current Protocols in Molecular Biology. John Wiley & Sons, New York. 1987.
- Stach JEM, Bathe S, Clapp JC, Burns RG. PCR-SSCP comparison of 16S rDNA sequence diversity in soil DNA obtained using different isolation and purification methods. FEMS Microbiol Ecol. 2001;36:139-151.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697-703.
- Msogoya T, Kanyagha H, Mutigitu J, Kulebelwa M, Mamiro D. Identification and management of microbial contaminants of banana in vitro cultures. J Appl Biosci. 2012;55:3987-3994.
- Gomez KA, Gomez AA. Statistical Procedure for Agricultural Research, 2nd edn. John Wiley and Sons, New York. 1984.
- Elshafie A, Alkindi AY, Al-Busaidi S, Bakheit C, Albahry SN. Biodegradation of crude oil and n-alkanes by fungi isolated from Oman. Mar Pollut Bull. 2007;54:1692-1696.
- Perera M, Wijayarthna D, Wijesundera S, Chinthaka M, Seneviratne G, Jayasena S. Biofilm mediated synergistic degradation of hexadecane by a naturally formed community comprising Aspergillus flavus complex and Bacillus cereus group. BMC Microbiol. 2019;19:84.
- Dörr J, Hurek T, Reinhold-Hurek B. Type IV Pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol. 1998;30:7-17.
- Seneviratne G, Tennakoon NS, Weerasekara MLMAW, Nandasena KA. Polyethylene biodegradation by a developed Penicillium–Bacillus biofilm. Current Sci. 2006;90:20-21.
- Benoit I, Esker MH, Patyshakuliyeva A, Mattern DJ, Blei F, Zhou M, et al. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ Microbiol. 2015;17:2099–2113.
- Arora DK, Gupta S. Effect of different environmental conditions on bacterial chemotaxis toward fungal spores. Can J Microbiol. 1993;39:922-931.
- Gaballa A, Abeysinghe PD, Urich G, Matthijs S, De Greve H, Cornelis P, et al. Trehalose induces antagonism towards Pythium debaryanum in Pseudomonas fluorescens ATCC 17400. Appl Environ Microbiol. 1997;63:4340–4345.
Citation: Utami D, Widianto D, Rohman MS, Satriyo HH, Sheila, Anggun J, et al. (2019) Synergistic Capability of Bacterial-fungal Coculture to Degrade Drill Cutting Hydrocarbon. J Microb Biochem Technol. 11:425. doi: 10.35248/1948-5948.19.11.425
Copyright: © 2019 Utami D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.