Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 11, Issue 9
Studies on Predominant Epiphytic Micro-flora as Antagonists to Postharvest Pathogens of Apple
Aqleema Banoo*, Efath Shahnaz, Saba Banday, Rovidha Rasool, Taibah Bashir and Rabia LatifReceived: 15-Jul-2020 Published: 08-Sep-2020, DOI: 10.35248/2157-7471.20.11.512
Abstract
Apple (Malus domestica Borkh) is an important horticultural crop that is affected by the number of diseases round the year. The fruit is particularly susceptible to a number of pathogens both pre- and post-harvest. Management of these diseases is based mostly on the application of synthetic fungicides with obvious disadvantages of environmental pollution, health hazards, pathogen resistance, etc. In the present study, eleven epiphytes were isolated using potato dextrose agar, nutrient agar and yeast maltose agar media. Amongst them, five fungal isolates viz., Aspergillus sp. (I1), Penicillium sp. (I2), Fusarium sp. (I3), Rhizopus sp. (I4) and Alternaria sp. (I5) and six bacterial isolates viz., Pseudomonas sp. (I6), Pseudomonas sp. (I7), Bacillus sp. (I8), Bacillus sp. (I9), Staphylococcus sp. (I10) and Micrococcus sp. (I11) were predominantly noticed under all the three methods (leaf impression, serial dilution and fruit washing) and were hence, used for further studies. The highest average colony count of 3.62 colonies/cm2 was recorded in fruit washing method followed by leaf impression (3.17) and lowest in serial dilution method (2.12). The in vitro screening of various bacterial and fungal epiphytes revealed that isolates of Pseudomonas sp. (I6) and Bacillus (I8 and I9) were the only bacterial strains capable of inhibiting the growth of all the test pathogens using dual culture method. Assays on wounded apples revealed that Pseudomonas sp. I6 at 107 cfu/ml was effective antagonist against Penicillium sp. and Fusarium sp., Bacillus sp. I9 at 107 cfu/ml was effective antagonist against Alternaria sp., whereas, Bacillus sp. I8 at 107 cfu/ml was most effective antagonist against Diplodia sp. The present study revealed that the antagonists were more or less efficient towards each pathogen and can be utilized for the management of post-harvest diseases of apple.
Keywords
Apple; Epiphytes; Biological control; Antagonism
Introduction
Apple, being a perishable commodity is subject to qualitative as well as quantitative pre- and post-harvest losses. Fungal pathogens are able to incite greater damage owing to the favourable fruit storage conditions and low resistance of the plants [1]. The important fungal pathogens responsible for causing major economic losses during postharvest storage conditions include Alternaria sp, Aspergillus niger, Botrytis sp, Cephalothecium roseum, Colletotrichum sp, Fusarium radiciola, Glomerella rufomaculans, G. cingulata, Monilinia fructigena, Neofabraea funera, Pencillium digitatum, P. expansum and Rhizopus nigricans, etc. [2-5].
Management of pre- and post-harvest fungal infections is principally based on the use of synthetic fungicides because of their effectiveness and low cost. Many of the fungicides such as benzimidazole and dicarboximide fungicides that are still available for use are losing their effectiveness due to the development of resistance in many postharvest pathogens [6]. Moreover, postharvest use of fungicides has been reduced due to the development of pathogen resistance, the public concern about presence of fungicide residues in food, associated environmental hazards and the lack of replacement of fungicides [7]. Currently, substantial progress has been made in finding alternatives to synthetic post-harvest fungicides and several microbial bio-control agents have been reported to control postharvest decay of pome fruits [8-10].
The microorganisms that are naturally present on the surfaces of fruits and vegetables, known as epiphytes, can be used as antagonists for the management of several diseases [11,12]. Such type of studies has been conducted [13-15]. Janisiewicz et al. [16] reported the feasibility of using mixtures of bacterial and yeast antagonists for the control of P. expansum on apples and suggested several modes of action employed by these microorganisms. Although, microbial antagonists can be applied either before or after harvest, postharvest applications are more effective than pre harvest applications [17]. Microbial cultures are applied either as postharvest sprays or as dips in an antagonist’s solution [18]. In Jammu and Kashmir, a lot of work has been done on the post-harvest diseases of apple and their management [3,19] but studies on phylloplane micro-organisms as bio-control agents for postharvest disease management is lacking. Hence, keeping in view the above facts, the present investigation shall be undertaken with the following objectives: 1) To estimate predominant epiphytes from leaves and fruits of apple and 2) To evaluate most effective epiphytes against most prevalent postharvest pathogens of apple.
Materials and Methods
Estimation of predominant epiphytes from leaves and fruits of apple
Collection of leaf and fruit samples: For the estimation of epiphytes, an orchard at Harwan, which had not been treated with fungicides for the last five years was selected. A group of seven trees, centrally located in the apple orchard was sampled. Fifty leaves were collected randomly from each tree, bulked, composited and taken immediately to the laboratory in sterilized perforated polythene bags for further studies. By using leaf area meter (Systronics leaf area meter 211) surface area of each leaf was measured and the average leaf area was expressed in cm². Similarly, fifty healthy blemish-free apple fruits were collected randomly from each apple tree in sterilized perforated polythene bags and brought to the laboratory for further studies. Length and diameter of collected healthy fruits were measured with the help of a digital Vernier caliper and an average fruit length and diameter were expressed in centimeters (cm).
Isolation of epiphytes by leaf impression method: The composited and selected leaves were washed with sterilized 70 distilled water in order to remove dirt and dust and were air dried. Dorsal and ventral surface of each leaf was pressed momentarily against the surface of potato dextrose agar plates at three places separately. The plates were properly labeled and incubated in an inverted position at 24 ± 2°C for fungi and 26 ± 2°C for bacteria for 3 days. The similar process was repeated with nutrient agar media and yeast agar media plates. The emergence and development of colonies was counted after every 24 hours [20]. Colonies of bacteria, actinomycetes and fungi per centimeter square of leaf was calculated by using the formula:
Colonies/cm² =Total No. of colonies / Leaf area
Isolation of epiphytes by serial dilution method: Five discs, each of 5 mm diameter were cut from every leaf using a sterile 5 mm cork borer. A total of 250 leaf discs were obtained, transferred to 100 ml of sterilized distilled water and stirred for 20 minutes till a suspension was obtained. The contents were shaken for uniform distribution of the cell / spores and 0.1 ml of aliquots from 10-1 and 10-2 dilutions using a sterile pipette were transferred to each sterile plates containing potato dextrose agar, nutrient agar, yeast maltose agar medium using L-shaped spreader. The plates were properly labeled and incubated at 24 ± 2°C for fungi and 26 ± 2°C for bacteria in an inverted position. The development of colonies was monitored and counted after every 24 hours for three days. Microbial population/cm² was calculated by applying the formula [20].
Propagules/cm² = Total No. of spores in 0.1 ml × 100 / Total area of 250 discs × 2
(Area of one leaf disc = πr2, where r is the radius of the disc in cm)
Isolation of epiphytes by fruit washing method: For isolation of epiphytes from fruits, selected fruit were dipped individually in a container of 150 ml sterile water and shaken for 20 minutes on a mechanical shaker at 250 rpm, to break free the micro-organisms. Each fruit was washed twice. The contents were shaken for uniform distribution of the cell / spores and 0.1 ml of aliquots from 10-1 dilutions using a sterile pipette was transferred to sterile plates containing potato dextrose agar medium, yeast maltose agar medium and 10-2 dilution was transferred to nutrient agar medium. Same procedure was repeated for second washing. The plates were properly labelled and incubated at 24 ± 2°C for fungi and 26 ± 2°C for bacteria. The development of colonies was monitored and counted after every 24 hrs. Microbial population/cm² of fruit surface was calculated by applying the formula [20].
Propagules/cm² = Total No. of colonies in 0.1 ml × 150 / Total area of apple fruit surface
(Area of apple fruit surface = 4πr², where r is the radius of the disc in cm)
Identification: The fungal colonies obtained were studied for colony characters, such as colony colour, margin, aerial growth and microscopic observations with respect to nature of mycelium, spore bearing structure, spore shape etc. The features of all the isolates were compared with the description in the standard manuals [21,22].
The bacterial colonies were grown at 26 ± 2°C for 24 hours on specific media viz., NA and King’s B medium slants or plates. The colony morphology was studied on plates after streaking a loop full of isolated colony. The bacterial isolates were Gram stained and slides observed under Gaynor microscope at 100X magnification. Cell shape, size, Gram reaction was observed and photographed [23]. The bacterial cultures were examined for various morphological, biochemical and physiological characteristics as per the procedure described in Bergey’s manual of Determinative Bacteriology.
Evaluation of fungal/bacterial epiphytes for their antagonistic property
In vitro screening of bacterial and fungal epiphytes for antagonism: All bacterial and fungal isolates were screened in vitro for antagonism against the most important apple postharvest pathogens by dual culture method [24,25]. 5 mm disc of the 7 day old culture of the pathogen was placed in the center of the Petri plate (90 mm diameter) under controlled conditions. The challenging isolates were streaked as a spore or cell suspension (for bacteria) or placed as an agar plug (for fungi) at equidistant positions from rim of Petri plate, after 3 days of incubation of pathogen. Plates not inoculated with epiphyte served as control. The plates were placed in BOD incubator at 24 ± 2℃ for 10 days after which they were evaluated for antagonistic activity. Per cent growth inhibition was determined by the formula of Skidmore [26].
Growth inhibition = Kr – r1 / Kr × 100
Where Kr represents the distance (measured in mm) of fungal growth from the point of inoculation to the colony margin on control plates, r₁ the distance of the fungal growth from the point of inoculation to the colony margin in the direction of the antagonist. Per cent growth inhibition was categorized on a scale Korsten [27] from 0 to 4 i.e., 0% = 0, 1 to 25% = 1, 26-50% = 2, 51-75% = 3 and 76-100% = 4. Isolates that reduced pathogen development by producing a demarcation zone or growth inhibition were selected for subsequent evaluation of antagonism on apple.
Assays on wounded apples: The laboratory tests were carried out on healthy apples cv. Red Delicious that had not been treated with any fungicides, as per method described [28]. Apples were stored in cold storage at Division of Post-Harvest Technology, SKUAST-K, Shalimar, Srinagar, up to 5 weeks at 4°C and 95% relative humidity before being used. They were surface sterilized by soaking in 70 per cent ethanol for 3 minutes. As the pathogens can infect apples only through wounds or lenticels, four circular wounds were introduced through the peel of each set of three fruits using a cork borer 5 mm in diameter and 3 mm in depth from the surface. One wound of each apple fruit was treated with 30 μl of cell suspension of bacterial bio-agents at three different concentrations of 105, 106 and 107 cfu/ ml and the other three wounds on each apple were treated with carbendazim at 0.05 per cent, mancozeb at 0.3 per cent and water, respectively. The treatments carbendazim (0.05%) and mancozeb (0.3%) served as standard and water as check. The concentration of bacterial bioagents was determined by dilution plating technique [9]. After 10 minutes of treatment, 20 μl of different pathogens at 2 × 104 spores per ml concentration (using haemocytometer) were inoculated and kept at ambient room storage conditions (22 ± 2°C). The same procedure was repeated on other set of apples which were kept under controlled atmospheric conditions (4℃) with relative humidity (90-95%) in Division of Post-Harvest Technology, SKUAST-K, Shalimar, Srinagar. The whole experiment was laid under completely randomized block design (CRBD) with three replications.
Statistical analysis: The data collected was analysed statistically using analysis of variance technique [29]. The data was transformed wherever necessary using OP STAT and R- software [30] at 5 per cent level of probability.
Results
Isolation and Identification of epiphytes
Five fungal isolates of epiphytes viz., Aspergillus sp. (I1), Penicillium sp. (I2), Rhizopus sp. (I4), Fusarium sp. (I3), and Alternaria sp. (I5) were isolated from leaf and fruit surface using three different methods (Plate 1) and isolates were identified on the basis of their morphological characteristics which were compared with authenticated descriptions [21,22]. Six bacterial epiphytes viz., Pseudomonas sp. (I6 and I7), Bacillus sp. (I8 and I9), Staphylococcus sp. (I10) and Micrococcus sp. (I11) were isolated. For identification purposes, morphological and biochemical characters of these isolates were studied as per the procedures described [31] (Figure 1).
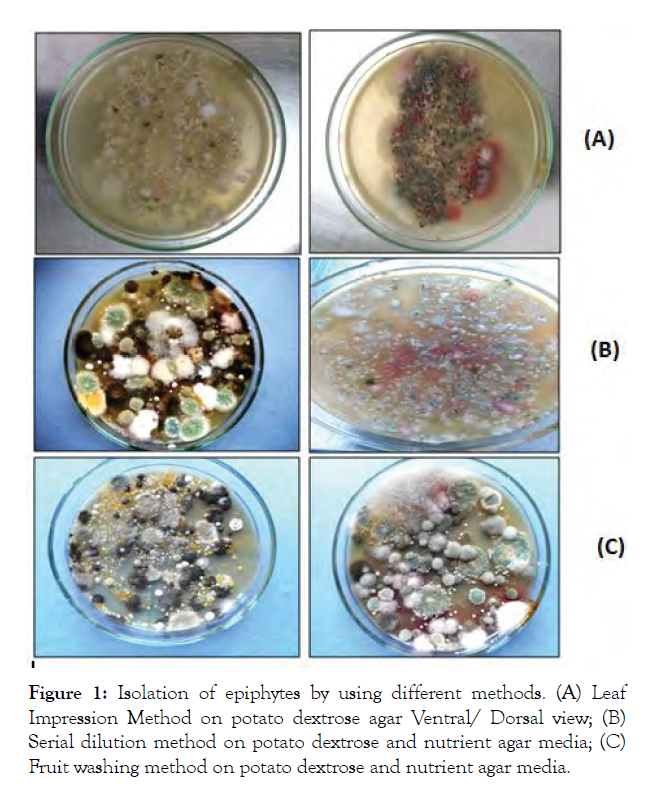
Figure 1: Isolation of epiphytes by using different methods. (A) Leaf Impression Method on potato dextrose agar Ventral/ Dorsal view; (B) Serial dilution method on potato dextrose and nutrient agar media; (C) Fruit washing method on potato dextrose and nutrient agar media.
Estimation of predominant epiphytes by leaf impression method
The mean populations (Table 1) of all microbial isolates viz., I1, I2, I3, I4, I7, I9, I10 and I12 on PDA (0.40 colonies per cm²) and YMA media (0.40 colonies/cm²) were significantly at par, but significantly different from NA media (0.34 colonies per cm²). However, a comparison of different media revealed that the microbial count of I9 (1.03 colonies per cm²) was highest on YMA, of I7 (0.95 colonies per cm²) on NA and of I10 (0.90 colonies per cm²) on PDA. The mean number of colonies was highest in case of I7 (0.92 colonies per cm²) and differed significantly from I9 (0.63 colonies per cm²) and others (0.53 colonies per cm²). The lowest mean numbers of colonies were recorded in case of I3, followed by I4 and I1 (0.02, 0.03 and 0.29 colonies per cm² respectively). The results are also shown in graph (Figure 2).
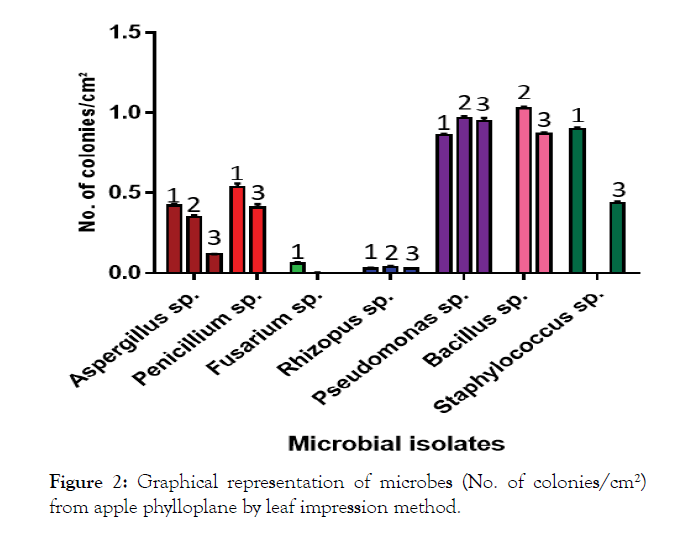
Figure 2: Graphical representation of microbes (No. of colonies/cm²) from apple phylloplane by leaf impression method.
| Microbial isolates | No. of colonies/cm² | ||||
|---|---|---|---|---|---|
| PDA Medium | YMA Medium | NA Medium | Overall mean | ||
| I1 | Aspergillus sp. | 0.42 (0.648) | 0.35 (0.591) | 0.12 (0.346) | 0.29 (0.538)c |
| I2 | Penicillium sp. | 0.54 (0.734) | 0.41 (0.460) | 0.00 (0.000) | 0.31 (0.556)d |
| I3 | Fusarium sp. | 0.06 (0.244) | 0.00 (0.000) | 0.00 (0.000) | 0.02 (0.414)a |
| I4 | Rhizopus sp. | 0.03 (0.173) | 0.04 (0.200) | 0.03 (0.173) | 0.03 (0.173)b |
| I7 | Pseudomonas sp. | 0.86 (0.927) | 0.97 (0.984) | 0.95 (0.974) | 0.92 (0.959)g |
| I9 | Bacillus sp. | 0.00 (0.000) | 1.03 (1.014) | 0.87 (0.932) | 0.63 (0.793)f |
| I10 | Staphylococcus sp. | 0.90 (0.948) | 0.00 (0.000) | 0.44 (0.663) | 0.44 (0.663)e |
| Mean | 0.40 (0.524)B | 0.40 (0.464)B | 0.34 (0.441)A | ||
Table 1: Microbial count of major epiphytes from apple phylloplane by leaf impression method.
Estimation of predominant epiphytes by serial dilution method
Perusal of the data presented in Table 2 revealed that the mean number of colonies of microbial isolates on PDA media was 0.20 per cm² which was significantly at par with the number of colonies on YMA (0.19 colonies per cm²) but significantly differed from NA (0.41 colonies per cm²). However, a comparison of different media revealed that the microbial count of I6 (0.92 colonies per cm²) was highest on NA media, of I6 (0.42 colonies per cm²) on PDA media and of I7 (0.40 colonies per cm²) on YMA media. The mean number of colonies significantly differed and was highest in case of I6 (0.55 colonies per cm²) followed by I7 (0.50 colonies per cm²) and I8 (0.40 colonies per cm²). The lowest mean numbers of colonies were recorded in case of I3 followed by I2 and I1 i.e. 0.02, 0.10 0.14 colonies per cm² of leaf. The results are also shown in graph (Figure 3).
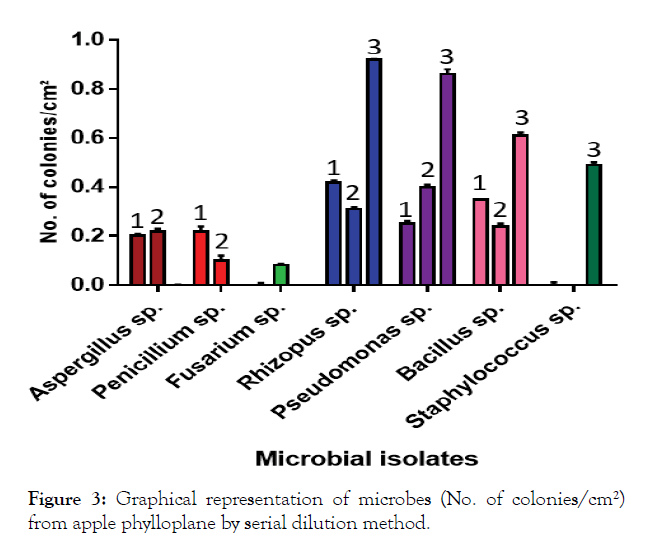
Figure 3: Graphical representation of microbes (No. of colonies/cm²) from apple phylloplane by serial dilution method.
| Microbial isolates | No. of colonies/cm² | ||||
|---|---|---|---|---|---|
| PDA Medium | YMA Medium | NA Medium | Overall mean | ||
| I1 | Aspergillus sp. | 0.20 (0.447) | 0.22 (0.469) | 0.0 (0.000) | 0.14 (0.374)c |
| I2 | Penicillium sp. | 0.22 (0.469) | 0.10 (0.316) | 0.0 (0.000) | 0.10 (0.316)b |
| I3 | Fusarium sp. | 0.0 (0.000) | 0.08 (0.282) | 0.0 (0.000) | 0.02 (0.141)a |
| I4 | Rhizopus sp. | 0.42 (0.648) | 0.31 (0.558) | 0.92 (0.951) | 0.55 (0.741)g |
| I7 | Pseudomonas sp. | 0.25 (0.500) | 0.40 (0.632) | 0.86 (0.927) | 0.50 (0.707)f |
| I9 | Bacillus sp. | 0.35 (0.591) | 0.24 (0.489) | 0.61 (0.781) | 0.40 (0.632)e |
| I10 | Staphylococcus sp. | 0.0 (0.000) | 0.0 (0.000) | 0.49 (0.700) | 0.16 (0.400)d |
| Mean | 0.20 (0.379)A | 0.19 (0.392)A | 0.41 (0.479)B | ||
Table 2: Microbial count of major epiphytes from apple phylloplane by serial dilution method.
Estimation of predominant epiphytes by fruit washing method
A perusal of the data presented in Table 3 revealed that the mean number of colonies of microbial isolates on PDA medium was 0.50 per cm². In case of YMA medium, the mean number of colonies of microbial isolates was 0.47 colonies per cm². While as, on NA medium, the mean number of colonies of microbial isolates was 0.44 colonies/cm² of leaf. However, a comparison of different media revealed that the microbial count of I8 (0.98 colonies per cm²) was highest on NA medium, I1 (0.91 colonies per cm²) on PDA medium and 0.81 colonies per cm² on YMA medium. The mean number of colonies significantly differed and were highest in case of I6 (0.86 colonies/cm²) followed by I9 (0.79 colonies per cm²) and I1 (0.57 colonies per cm²). The lowest number of colonies were recorded in case of I3 (0.03 colonies per cm²) followed by I2 (0.30 colonies per cm²) and I8 (0.32 colonies per cm²). The results are also shown in graph (Figure 4).
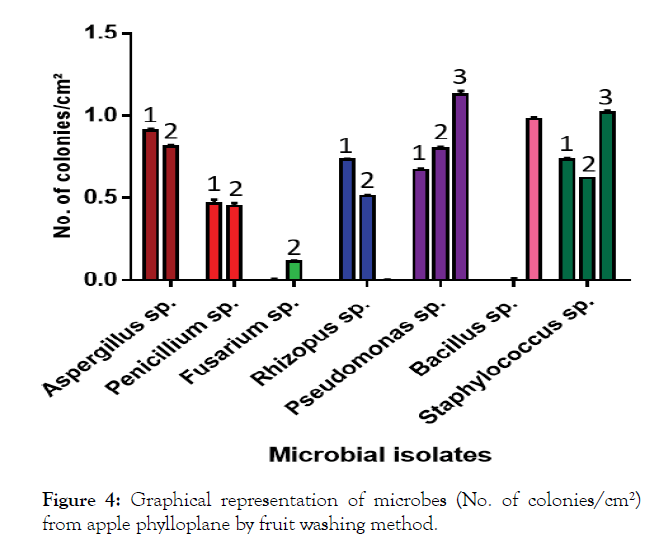
Figure 4: Graphical representation of microbes (No. of colonies/cm²) from apple phylloplane by fruit washing method.
| Microbial isolates | No. of colonies/cm² | ||||
|---|---|---|---|---|---|
| PDA Medium | YMA Medium | NA Medium | Overall mean | ||
| I1 | Aspergillus sp. | 0.91 (0.953) | 0.81 (0.900) | 0.0 (0.000) | 0.57 (0.754)e |
| I2 | Penicillium sp. | 0.47 (0.685) | 0.45 (0.670) | 0.0 (0.000) | 0.30 (0.547)b |
| I3 | Fusarium sp. | 0.0 (0.000) |
0.11 (0.331) | 0.0 (0.000) | 0.03 (0.173)a |
| I4 | Rhizopus sp. | 0.73 (0.854) | 0.51 (0.714) | 0.0 (0.000) | 0.41 (0.640)d |
| I7 | Pseudomonas sp. | 0.67 (0.818) | 0.80 (0.894) | 1.13 (1.060) | 0.86 (0.927)g |
| I9 | Bacillus sp. | 0.0 (0.000) |
0.0 (0.000) | 0.98 (0.989) | 0.32 (0.565)c |
| I10 | Staphylococcus sp. | 0.73 (0.854) | 0.62 (0.787) | 1.02 (1.009) | 0.79 (0.888)f |
| Mean | 0.50 (0.594)B | 0.47 (0.613)C | 0.44 (0.436)A | ||
CD (P ≤ 0.05) Isolate (I) = 0.003 Media (M) = 0.002 Isolate × Media = 0.006
Figures in brackets are square root transformed values.
Table 3: Microbial count of major epiphytes from apple fruits by fruit washing method.
Evaluation of fungal/bacterial epiphytes for their antagonistic property
In vitro screening of potential bacterial and fungal epiphytes for antagonism: All the isolates were screened against all the pathogens isolated from symptomatic apples (Penicillium, Fusarium, Alternaria and Diplodia sp.). Yeast maltose agar medium was used for this purpose, since it supported growth of fungi and bacteria both. A perusal of the data presented in Table 4 revealed that the six isolates viz., I3, I6, I7, I8, I9 and I10 inhibited Penicillium sp. 25 per cent (category 1 on a scale of 0-4), whereas, I1, I2, I4, I5 and I11 showed no inhibition in growth. The maximum growth inhibition (25-50%) in case of Alternaria sp. was recorded by I6 which was categorized in group 2 followed by I1, I2, I4, I8, I9, I10 and I11 (category 1). Fusarium sp. was inhibited up to 50 per cent by three isolates viz., I6, I8 and I9, whereas, Diplodia sp. was inhibited up to 50 per cent by only one isolate I8. These were placed in category 1 on 0-4 scale of Korsten. The microbial isolates I1, I4, I5, I7 and I8 inhibited Fusarium sp., whereas, I5, I6, I7, I9 and I10 inhibited Diplodia sp. up to 25 per cent (category 1). The data further revealed that under in vitro conditions, the growth of Penicillium sp. and Diplodia sp. were reduced by 6 isolates, whereas, Alternaria sp. and Fusarium sp. were inhibited by 8 microbial isolates. Of all microbial isolates evaluated, I6, I8 and I9 were the only bacterial strains capable of inhibiting the growth of all the tested pathogens in the category of 1 or 2 and thus proved most effective and were selected for subsequent evaluation of antagonism on apple (Figure 5).
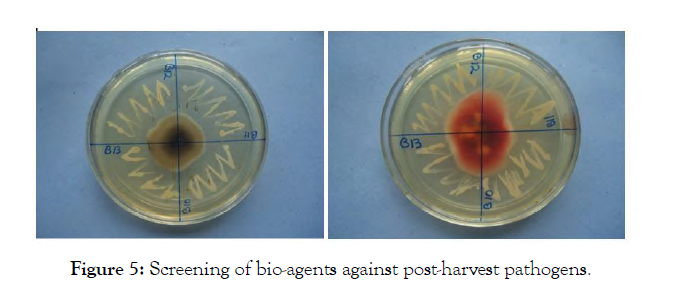
Figure 5: Screening of bio-agents against post-harvest pathogens.
| Microbial Isolates | Growth inhibition (GI) category | No. of Pathogens inhibited | ||||
|---|---|---|---|---|---|---|
| Penicillium sp. | Alternaria sp. | Fusarium sp. | Diplodia sp. | |||
| I1 | Aspergillus sp. | 0 | 1 | 1 | 0 | 2 |
| I2 | Penicillium sp. | 0 | 1 | 0 | 0 | 1 |
| I3 | Fusarium sp. | 1 | 0 | 0 | 0 | 1 |
| I4 | Rhizopus sp. | 0 | 1 | 1 | 0 | 2 |
| I5 | Alternaria sp. | 0 | 0 | 1 | 1 | 2 |
| I6 | Pseudomonas sp. | 1 | 2 | 2 | 1 | 4 |
| I7 | Pseudomonas sp. | 1 | 0 | 1 | 1 | 3 |
| I8 | Bacillus sp. | 1 | 1 | 2 | 2 | 4 |
| I9 | Bacillus sp. | 1 | 1 | 2 | 1 | 4 |
| I10 | Staphylococcus sp. | 1 | 1 | 0 | 1 | 3 |
| I11 | Micrococcus sp. | 0 | 1 | 1 | 0 | 2 |
| Total inhibitory isolates | 6 | 8 | 8 | 6 | ||
Percent growth inhibition was determined after 7 days by the formula of Skidmore.
Values were categorized on a scale from 0 to 4, where 0= No growth inhibition 1 = 1% to 25 %, 2 = 26% to 50 %, 3 = 51% to 75% and 4 = 76% to 100%.
Table 4: Screening of potential bio-control agents against important postharvest apple pathogens.
Evaluation of most effective epiphytes against prevalent postharvest pathogens of apple: From the in vitro tests, three biocontrol agents were selected for their efficacy to reduce lesion development. The bio-control agents at different concentrations (105, 106and 107cfu / ml) were tested against the pathogen on artificially wounded apples along with carbendazim (0.05%) and mancozeb (0.3%) as standard and water as check. These wounded apples were subsequently stored under ambient room storage conditions (S1) and controlled atmospheric conditions at 4°C (S2).
Assays on wounded apples against Penicillium sp.: A perusal of the data presented in the Table 5 revealed that all the isolates significantly reduced the lesion development of Penicillium sp. on apple. Moreover, perusal of data also revealed that under both storage conditions B1C1 (I6 at 107 cfu/ml) proved most effective antagonistic against Penicillum sp. by providing only 3.1 mm lesion development followed by B1C2 (I6 at 106 cfu/ml) providing 4.6 mm lesion development and lowest lesion development of 6.3 mm was by B2C3 (I8 at 105 cfu/ml) and B3C3 (I9 at 105 cfu/ml) respectively.
| S. No. | Treatments | Ambient condition (S1) | Sub-Mean | Controlled condition (S2) |
Sub-Mean | 10 D | 15 D | 20 D | Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 D | 15 D | 20 D | 10 D | 15 D | 20 D | ||||||||
| 1 | B1C1 | 0.0 (1.000) |
5.7 (2.588) |
7.9 (2.983) |
4.5 (2.191) |
0.0 (1.000) |
0.0 (1.000) |
5.4 (2.530) |
1.8 (1.510) |
0.0 (1.000) |
2.8 (1.794) |
6.6 (2.757) |
3.1 (1.850)b |
| 2 | B1C2 | 5.5 (2.556) |
7.0 (2.828) |
8.6 (3.098) |
7.0 (2.828) |
0.0 (1.000) |
0.0 (1.000) |
6.3 (2.702) |
2.1 (1.567) |
2.7 (1.778) |
3.5 (1.914) |
4.9 (2.900) |
4.5 (2.197)e |
| 3 | B1C3 | 5.7 (2.588) |
7.2 (2.864) |
8.8 (3.130) |
7.2 (2.861) |
0.0 (1.000) |
5.5 (2.550) |
7.0 (2.828) |
4.1 (2.126) |
2.8 (1.794) |
6.3 (2.707) |
7.9 (2.979) |
5.6 (2.493)g |
| 4 | B2C1 | 6.5 (2.739) |
7.0 (2.828) |
8.7 (3.114) |
7.4 (2.894) |
0.0 (1.000) |
0.0 (1.000) |
6.0 (2.646) |
2.0 (1.549) |
3.2 (1.869) |
3.5 (1.914) |
7.3 (2.880) |
4.7 (2.221)f |
| 5 | B2C2 | 6.5 (2.739) |
7.7 (2.950) |
9.2 (3.194) |
7.8 (2.961) |
0.0 (1.000) |
6.5 (2.739) |
7.5 (2.915) |
4.6 (2.218) |
3.2 (1.869) |
7.2 (2.844) |
8.3 (3.055) |
6.2 (2.589)i |
| 6 | B2C3 | 6.7 (2.775) |
7.7 (2.950) |
9.4 (3.225) |
7.9 (2.983) |
0.0 (1.000) |
6.7 (2.775) |
7.5 (2.915) |
4.7 (2.230) |
3.3 (1.887) |
7.2 (2.862) |
8.4 (3.070) |
6.3 (2.598)kj |
| 7 | B3C1 | 5.0 (2.449) |
6.5 (2.739) |
8.3 (3.050) |
6.6 (2.746) |
0.0 (1.000) |
0.0 (1.000) |
6.3 (2.702) |
2.1 (1.567) |
2.5 (1.725) |
3.2 (1.869) |
7.3 (2.876) |
4.3 (2.157)d |
| 8 | B3C2 | 6.3 (2.702) |
7.8 (2.966) |
9.1 (3.178) |
7.7 (2.949) |
0.0 (1.000) |
6.0 (2.646) |
7.0 (2.828) |
4.3 (2.158) |
3.1 (1.851) |
6.9 (2.806) |
8.0 (3.003) |
6.0 (2.553)h |
| 9 | B3C3 | 6.4 (2.720) |
8.2 (3.033) |
10.3 (3.362) |
8.3 (3.038) |
0.0 (1.000) |
6.0 (2.646) |
7.0 (2.828) |
4.3 (2.158) |
3.2 (1.860) |
7.1 (2.839) |
8.6 (3.095) |
6.3 (2.598)j |
| 10 | Carbendazim 50WP (0.05%) | 0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000)a |
| 11 | Mencozeb 75WP (0.3%) | 0.0 (1.000) |
7.7 (2.950) |
8.4 (3.066) |
5.3 (2.339) |
0.0 (1.000) |
0.0 (1.000) |
6.0 (2.646) |
2.0 (1.549) |
0.0 (1.000) |
3.8 (1.975) |
7.2 (2.856) |
3.6 (1.944)c |
| 12 | Water | 9.0 (3.162) |
18.6 (4.427) |
25.0 (5.099) |
17.5 (4.229) |
0.0 (1.000) |
7.7 (2.950) |
13.0 (3.742) |
6.9 (5.564) |
4.5 (2.081) |
13.2 (3.688) |
19.0 (4.420) |
12.2 (3.397)k |
| Mean | 4.8 (2.286) |
7.5 (2.844) |
9.4 (3.125) |
7.2 (2.751)B |
0.0 (1.000) |
3.2 (1.859) |
6.5 (2.690) |
2.2 (1.850)A |
2.4 (1.643)A |
5.3 (2.351)B |
7.8 (2.908)C |
||
CD (p<0.05)
Treatments (T) = 0.003 T × S = 0.0004 B1: Pseudomonas sp. C1: Concentration (107cfu/ml) S1: Ambient room condition
Storage (S) = 0.001 T × D = 0.005 B2: Bacillus sp. C2: Concentration (106 cfu/ml) S2: Controlled room condition
Duration (D) = 0.002 S × D = 0.002 B3: Bacillus sp. C3: Concentration (105cfu/ml)
T × S × D = 0.008. Figures in brackets are square root transformed values.
Table 5: Effect of different concentrations of antagonistic bacterial isolates on the reduction of lesion development (mm) of Penicillium sp. Under different storage conditions.
However, S2 was significantly different from S1 and proved better storage condition in reduction of lesion development by pathogens with a mean of (2.2 mm) in comparison to S1 (7.2 mm). By comparing isolates with different checks, carbandazim 0.05% proved to be most effective as there was no lesion development and water proved to be least effective by providing highest lesion development in both S1 and S2.
Assays on wounded apples against Alternaria sp.: The data presented in the Table 6 revealed that both storage conditions B3C1 (I9 at 107 cfu/ml) proved effective antagonistic against Alternaria sp. by providing only 3 mm lesion development followed by B1C1 (I6 at 107 cfu/ml) providing 3.2 mm lesion development and lowest lesion development of 5.4 mm was by B1C3 (I6 at 105 cfu/ml) and B3C3 (I9 at 105 cfu/ml) respectively. However, S2 was significantly different from S1 and proved better storage condition in reduction of lesion development by pathogens with a mean of (3.2 mm) in comparison to S1 (5.6 mm). By comparing isolates with different checks, carbendazim 0.05% proved to be most effective as there was no lesion development and water proved to be least effective by providing highest lesion development in both S1 and S2.
| S. No. | Treatments | Ambient condition (S1) | Sub-Mean | Controlled condition (S2) |
Sub-Mean | 10 D | 15 D | 20 D | Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B1C1 | 0.0 (1.000) |
6.2 (2.683) |
7.9 (2.983) |
4.7 (2.222) |
0.0 (1.000) |
0.0 (1.000) |
5.2 (2.490) |
1.7 (1.497) |
0.0 (1.000) |
3.1 (1.842) |
6.5 (2.737) |
3.2 (1.859)c |
| 2 | B1C2 | 5.6 (2.569) |
6.7 (2.775) |
8.2 (3.033) |
6.8 (2.792) |
0.0 (1.000) |
0.0 (1.000) |
5.4 (2.530) |
1.8 (1.510) |
2.8 (1.784) |
3.3 (1.887) |
6.8 (2.781) |
4.3 (2.151)d |
| 3 | B1C3 | 5.8 (2.608) |
7.1 (2.846) |
8.8 (3.130) |
7.2 (2.861) |
0.0 (1.000) |
0.0 (1.000) |
6.0 (2.646) |
2.0 (1.549) |
2.9 (1.804) |
3.5 (1.923) |
7.4 (2.888) |
4.6 (2.205)e |
| 4 | B2C1 | 6.4 (2.720) |
6.9 (2.811) |
8.0 (3.000) |
7.7 (2.844) |
0.0 (1.000) |
5.2 (2.490) |
6.2 (2.683) |
3.8 (2.058) |
3.2 (1.860) |
6.0 (2.650) |
7.1 (2.842) |
5.4 (2.451)g |
| 5 | B2C2 | 7.1 (2.846) |
7.6 (2.933) |
8.4 (3.066) |
7.7 (2.948) |
0.0 (1.000) |
5.4 (2.530) |
6.6 (2.757) |
4.0 (2.096) |
3.5 (1.923) |
6.5 (2.731) |
7.5 (2.911) |
5.8 (2.522)h |
| 6 | B2C3 | 7.1 (2.846) |
7.7 (2.950) |
8.5 (3.082) |
7.7 (2.959) |
0.0 (1.000) |
5.4 (2.530) |
7.0 (2.828) |
4.1 (2.119) |
3.2 (1.923) |
6.5 (2.740) |
7.7 (2.955) |
5.8 (2.522)h |
| 7 | B3C1 | 0.0 (1.000) |
5.6 (2.569) |
6.9 (2.811) |
4.1 (2.127) |
0.0 (1.000) |
0.0 (1.000) |
5.7 (2.588) |
1.9 (1.529) |
0.0 (1.000) |
2.8 (1.785) |
6.3 (2.700) |
3.0 (1.828)b |
| 8 | B3C2 | 0.0 (1.000) |
5.8 (2.608) |
7.4 (2.898) |
4.4 (2.169) |
0.0 (1.000) |
0.0 (1.000) |
6.2 (2.683) |
2.0 (1.561) |
0.0 (1.000) |
2.9 (1.804) |
6.8 (2.791) |
3.2 (1.859)c |
| 9 | B3C3 | 5.6 (2.569) |
6.3 (2.702) |
8.3 (3.050) |
6.7 (2.773) |
0.0 (1.000) |
5.2 (2.490) |
6.4 (2.720) |
3.8 (2.070) |
2.8 (1.785) |
5.7 (2.596) |
7.3 (2.885) |
5.2 (2.422)f |
| 10 | Carbendazim 50WP (0.05%) | 0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000)a |
| 11 | Mencozeb 75WP (0.3%) | 0.0 (1.000) |
5.6 (2.569) |
7.0 (2.828) |
4.2 (2.132) |
0.0 (1.000) |
0.0 (1.000) |
5.9 (2.627) |
1.9 (1.542) |
0.0 (1.000) |
2.8 (1.785) |
6.4 (2.728) |
3.0 (1.828)b |
| 12 | Water | 8.2 (3.033) |
13.9 (3.860) |
20.3 (4.615) |
14.1 (3.836) |
0.0 (1.000) |
5.9 (2.627) |
9.6 (3.356) |
5.1 (2.294) |
4.1 (2.017) |
9.9 (3.243) |
14.9 (3.935) |
9.6 (3.065)i |
| Mean | 3.8 (2.016) |
6.6 (2.692) |
8.3 (2.958) |
6.2 (2.555)B |
0.0 (1.000) |
2.2 (1.639) |
5.8 (2.567) |
2.6 (1.735)A |
1.9 (1.508)A |
4.4 (2.165)B |
7.0 (2.763)C |
||
CD (p<0.05)
Treatments (T) = 0.002 T × S = 0.0002 B1: Pseudomonas sp. C1: Concentration (107cfu/ml) S1: Ambient room condition
Storage (S) = 0.001 T × D = 0.003 B2: Bacillus sp. C2: Concentration (106 cfu/ml) S2: Controlled room condition
Duration (D) = 0.001 S × D = 0.001 B3: Bacillus sp. C3: Concentration (105cfu/ml)
T × S × D = 0.004
Figures in brackets are square root transformed values.
Table 6: The effect of different concentration of antagonistic bacterial isolates on the reduction of lesion development (mm) Alternaria sp. under different storage conditions.
Assays on wounded apples against Diplodia sp.: A presual of data present in the Table 7 revealed that all the isolates significantly reduced the lesion development of Diplodia sp. on apple. Moreover, data also revealed that under both storage conditions B3C1 (I9 at 107 cfu/ml) proved most effective antagonistic against Diplodia sp. by providing only 1.9 mm lesion development followed by B1C1 (I6 at 107 cfu/ml) providing 2.7 mm lesion development and lowest lesion development of 5.8 mm was by B2C2 (I8 at 106 cfu/ml) and B2C3 (I8 at 105 cfu/ml) respectively. However, S2 was significantly different from S1 and proved better storage condition in reduction of lesion development by pathogens with a mean of (2.6 mm) in comparison to S1 (6.2 mm).
| S. No. | Treatments | Ambient condition (S1) | Sub-Mean | Controlled condition (S2) |
Sub-Mean | 10 D | 15 D | 20 D | Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B1C1 | 0.0 (1.000) |
5.3 (2.510) |
6.0 (2.646) |
3.7 (2.052) |
0.0 (1.000) |
0.0 (1.000) |
5.3 (2.510) |
1.8 (1.503) |
0.0 (1.000) |
2.6 (1.755) |
5.6 (2.578) |
2.7 (1.778)c |
| 2 | B1C2 | 5.3 (2.510) |
6.5 (2.739) |
9.0 (3.162) |
6.9 (2.804) |
0.0 (1.000) |
5.3 (2.510) |
6.1 (2.665) |
3.8 (2.058) |
2.6 (1.755) |
5.9 (2.624) |
7.5 (2.913) |
5.3 (2.431)h |
| 3 | B1C3 | 5.4 (2.530) |
6.8 (2.793) |
9.0 (3.162) |
7.0 (2.828) |
0.0 (1.000) |
5.3 (2.510) |
6.4 (2.720) |
3.9 (2.077) |
2.7 (1.765) |
6.0 (2.651) |
7.7 (2.941) |
5.4 (2.453)j |
| 4 | B2C1 | 0.0 (1.000) |
5.5 (2.550) |
6.7 (2.775) |
4.0 (2.108) |
0.0 (1.000) |
5.3 (2.510) |
5.8 (2.608) |
3.7 (2.039) |
0.0 (1.000) |
5.4 (2.530) |
6.2 (2.691) |
3.8 (2.074)e |
| 5 | B2C2 | 5.5 (2.549) |
6.3 (2.702) |
6.6 (2.757) |
6.1 (2.669) |
0.0 (1.000) |
5.8 (2.608) |
5.8 (2.608) |
3.8 (2.072) |
2.7 (1.775) |
6.0 (2.655) |
6.2 (2.682) |
4.9 (2.371)f |
| 6 | B2C3 | 5.7 (2.588) |
7.0 (2.828) |
7.7 (2.950) |
6.8 (2.789) |
0.0 (1.000) |
5.9 (2.627) |
6.2 (2.683) |
4.0 (2.103) |
2.8 (1.794) |
6.4 (2.728) |
6.9 (2.816) |
5.3 (2.431)i |
| 7 | B3C1 | 0 (1.000) |
0.0 (1.000) |
6.0 (2.646) |
2.0 (1.549) |
0.0 (1.000) |
0.0 (1.000) |
5.5 (2.550) |
1.8 (1.517) |
0.0 (1.000) |
0.0 (1.000) |
5.7 (2.598) |
1.9 (1.533)b |
| 8 | B3C2 | 5.4 (2.530) |
6.6 (2.757) |
8.0 (3.000) |
6.6 (2.762) |
0.0 (1.000) |
5.5 (2.550) |
6.3 (2.702) |
3.9 (2.084) |
2.7 (1.765) |
6.0 (2.653) |
7.1 (2.851) |
5.2 (2.423)g |
| 9 | B3C3 | 5.7 (2.588) |
7.0 (2.828) |
8.1 (3.017) |
6.9 (2.811) |
0.0 (1.000) |
5.7 (2.588) |
6.5 (2.744) |
4.0 (2.111) |
2.8 (1.794) |
6.3 (2.708) |
7.3 (2.881) |
5.4 (2.453)j |
| 10 | Carbendazim 50WP (0.05%) | 0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000)a |
| 11 | Mencozeb 75WP (0.3%) | 0.0 (1.000) |
5.7 (2.588) |
7.7 (2.950) |
4.4 (2.179) |
0.0 (1.000) |
0.0 (1.000) |
5.3 (2.510) |
1.7 (1.503) |
0.0 (1.000) |
2.8 (1.794) |
6.5 (2.730) |
3.1 (1.841)d |
| 12 | Water | 8.2 (3.033) |
13.4 (3.795) |
19.1 (4.483) |
13.5 (3.770) |
0.0 (1.000) |
9.6 (3.256) |
10.3 (3.362) |
6.6 (2.539) |
4.1 (2.017) |
11.5 (3.525) |
14.7 (3.922) |
10.1 (3.155)k |
| Mean | 3.4 (1.944) |
5.8 (2.507) |
7.8 (2.879) |
5.6 (2.443)B |
0.0 (1.000) |
4.0 (2.097) |
5.7 (2.555) |
3.2 (1.884A) |
1.7 (1.472) A |
4.9 (2.302)B |
6.7 (2.717) C |
||
CD (p<0.05)
Treatments (T) = 0.004 T × S = 0.0006 B1: Pseudomonas sp. C1: Concentration (107cfu/ml) S1: Ambient room condition
Storage (S) = 0.002 T × D = 0.008 B2: Bacillus sp. C2: Concentration (106 cfu/ml) S2: Controlled room condition
Duration (D) = 0.002 S × D = 0.003 B3: Bacillus sp. C3: Concentration (105cfu/ml)
T × S × D = 0.001
Figures in brackets are square root transformed values.
Table 7: Effect of different concentrations of antagonistic bacterial isolates on the reduction of lesion development (mm) of Diplodia sp. under different storage conditions.
Assays on wounded apples against Fusarium sp.: The data present in the Table 8 revealed that all the isolates significantly reduced the lesion development of Fusarium sp. on apple. However, S2 was significantly different from S1 and proved better storage condition in reduction of lesion development by pathogens with a mean of (2.7 mm) in comparison to S1 (5.6 mm). By comparing isolates with different checks, carbendazim 0.05% proved to be most effective as there was no lesion development and water proved to be least effective by providing highest lesion development in both S1 and S2.
| S. No. | Treatments | Ambient condition (S1) | Sub-Mean | Controlled condition (S2) |
Sub-Mean | 10 D | 15 D | 20 D | Mean | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B1C1 | 0.0 (1.000) |
5.3 (2.510) |
6.0 (2.646) |
3.7 (2.052) |
0.0 (1.00) |
0.0 (1.000) |
5.0 (2.449) |
1.6 (1.483) |
0.0 (1.000) |
2.6 (1.755) |
5.5 (2.548) |
2.6 (1.768)b |
| 2 | B1C2 | 0.0 (1.000) |
5.7 (2.588) |
6.3 (2.702) |
4.0 (2.097) |
0.0 (1.00) |
0.0 (1.000) |
5.0 (2.449) |
1.6 (1.483) |
0.0 (1.000) |
2.8 (1.794) |
5.6 (2.576) |
2.8 (1.790)c |
| 3 | B1C3 | 5.1 (2.470) |
5.8 (2.608) |
6.5 (2.739) |
5.8 (2.605) |
0.0 (1.00) |
0.0 (1.000) |
5.6 (2.569) |
1.8 (1.523) |
2.5 (1.735) |
2.9 (1.804) |
6.0 (2.654) |
3.8 (2.064) d |
| 4 | B2C1 | 0.0 (1.000) |
5.5 (2.549) |
6.8 (2.793) |
4.1 (2.114) |
0.0 (1.00) |
5.2 (2.490) |
5.4 (2.530) |
3.5 (2.007) |
0.0 (1.000) |
5.3 (2.520) |
6.1 (2.661) |
3.8 (2.060)d |
| 5 | B2C2 | 5.3 (2.510) |
5.8 (2.608) |
7.0 (2.828) |
6.0 (2.649) |
0.0 (1.00) |
5.4 (2.530) |
5.7 (2.588) |
3.7 (2.039) |
2.6 (1.755) |
5.6 (2.569) |
6.3 (2.708) |
4.8 (2.344)g |
| 6 | B2C3 | 5.4 (2.543) |
6.0 (2.646) |
7.3 (2.881) |
6.2 (2.690) |
0.0 (1.00) |
5.4 (2.530) |
5.6 (2.569) |
3.6 (2.033) |
2.7 (1.771) |
5.7 (2.588) |
6.4 (2.725) |
4.9 (2.361)h |
| 7 | B3C1 | 5.3 (2.510) |
5.6 (2.575) |
7.2 (2.865) |
6.0 (2.650) |
0.0 (1.00) |
5.0 (2.449) |
5.3 (2.510) |
3.4 (1.986) |
2.6 (1.755) |
5.3 (2.512) |
6.2 (2.688) |
4.7 (2.318)f |
| 8 | B3C2 | 5.3 (2.510) |
5.8 (2.620) |
8.2 (3.033) |
6.4 (2.721) |
0.0 (1.00) |
5.2 (2.490) |
5.5 (2.550) |
3.5 (2.013) |
2.6 (1.755) |
5.5 (2.555) |
6.8 (2.791) |
4.9 (2.361)h |
| 9 | B3C3 | 5.4 (2.543) |
6.3 (2.702) |
8.6 (3.098) |
6.7 (2.781) |
0.0 (1.00) |
5.4 (2.530) |
5.7 (2.588) |
3.7 (2.039) |
2.7 (1.771) |
5.8 (2.616) |
7.1 (2.843) |
5.2 (2.410)i |
| 10 | Carbendazim 50WP (0.05%) | 0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.00) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000) |
0.0 (1.000)a |
| 11 | Mencozeb 75WP (0.3%) | 5.1 (2.477) |
6.2 (2.689) |
7.7 (2.950) |
6.3 (2.705) |
0.0 (1.00) |
0.0 (1.000) |
5.3 (2.510) |
1.7 (1.503) |
2.5 (1.738) |
3.1 (1.845) |
6.5 (2.730) |
4.0 (2.104)e |
| 12 | Water | 7.6 (2.933) |
13.0 (3.742) |
18.0 (4.359) |
10.3 (3.678) |
0.0 (1.00) |
5.8 (2.608) |
9.0 (3.162) |
4.9 (2.257) |
3.8 (1.966) |
9.4 (3.175) |
13.5 (3.761) |
7.6 (2.967)j |
| Mean | 3.7 (2.041) |
5.9 (2.570) |
7.4 (2.824) |
5.6 (2.478)B |
0.0 (1.000) |
3.1 (1.886) |
5.2 (2.456) |
2.7 (1.781)A |
1.8 (1.521)A |
4.5 (2.228)B |
6.3 (2.640)C |
||
CD (p<0.05)
Treatments (T) = 0.018 T × S = 0.026B1: Pseudomonas sp. C1: Concentration (107cfu/ml) S1: Ambient room condition
Storage (S) = 0.008 T × D = 0.032 B2: Bacillus sp. C2: Concentration (106 cfu/ml) S2: Controlled room condition
Duration (D) = 0.009 S × D = 0.013 B3: Bacillus sp. C3: Concentration (105cfu/ml)
T × S × D = 0.044
Figures in brackets are square root transformed values.
Table 8: Effect of different concentrations of antagonistic bacterial isolates on the reduction of lesion development (mm) of Fusarium sp. under different storage conditions.
Discussion
There are very few studies on the epiphytes of apple and their role in the management of postharvest diseases of apple. The present investigation was, therefore, aimed at isolation and estimation of predominant epiphytes and screening of most effective antagonistic epiphytes against the post-harvest pathogens of apple cv. Red Delicious. This is the first report on epiphytes to be used as bioagent and thus can be incorporated in management strategies for apple diseases.
Five fungal epiphytes viz., Aspergillus sp. (I1), Penicillium sp. (I2), Fusarium sp. (I3), Rhizopus sp. (I4) and Alternaria sp. (I5) and six bacterial epiphytes viz., Pseudomonas sp. (I6), Pseudomonas sp. (I7), Bacillus sp. (I8), Bacillus sp. (I9), Staphylococcus sp. (I10) and Micrococcus sp. (I11) were isolated using different techniques viz., leaf impression, serial dilution and fruit washing methods on three different media (potato dextrose, nutrient agar and yeast agar media). The various methods used in this study to quantify the number of propagules/colonies of microbes are only a rough means of estimating the presence of micro-organisms on leaves [25]. It takes into account only those microbes which can be washed away from the leaves and grown on media. Similarly, Ogwu and Osawaru [32] estimated microfloral isolates on healthy leaves of mature okra and examined eight genera of fungi (Rhodotorula, Saccharomyces, Mucor, Trichothecium, Cladosporium, Rhizopus Aspergillus and Penicillium) and six genera of bacteria (Bacillus, Pseudomonas, Micrococcus, Proteus, Staphyloccocus, Serratia).
From all the three epiphytes isolation methods highest number of microbial colonies observed was Bacillus followed by Pseudomonas. This investigation shows that apple phylloplane is inhabited by various fungal and bacterial epiphytes. Most predominant microflora were Bacillus and Pseudomonas sp. This is in conformity with Bryk et al. [33] who isolated two strains of Pseudomonas sp. (B194 and B224) from apple leaves.
After screening the microbial isolates against all the pathogens, it has been observed that Pseudomonas sp. (I6), Bacillus sp. (I8) and Bacillus sp. (I9) were the only bacterial strains capable of inhibiting the radial mycelial growth of all four postharvest pathogens of apple, though at different inhibition levels. More or less, similar work was done by Grahovac et al. [34], who evaluated in vitro antifungal efficiency of four microorganisms viz., Streptomyces hygroscopicus, Saccharomyces cervisae, Bacillus cereus and Leuconostoc mesenteroides against apple fruit rot causing pathogens viz., Colletotrichum acutatum, C. gloeosporioides and F. avenaceum. In another study with P. expansum it was shown that Saccharomyces cervisae has good antagonistic activity and the strain was proposed to be used as a commercial biocontrol agent for storage apple [35]. The Bacillus subtilis and Trichoderma pseudokonongii were the first antagonists used for biological control of brown rot of stone fruits and gray mold of apple caused by Botrytis cinerea [36,37]. Sobiczewski [12] found Pseudomonas sp. as effective antagonist among the epiphytic microflora of apple.
Three most effective antagonistic epiphytes which showed maximum inhibition were then assessed against the four predominant pathogens for inhibiting lesion development on inoculated fruits. Assays on wounded apples revealed that Pseudomonas sp. I6 at 107 cfu/ml was effective antagonist against Penicillium sp. and Fusarium sp. Bacillus sp. I9 at 107 cfu/ml proved to be effective antagonist against Alternaria sp. Bacillus sp. I8 at 107 cfu/ml was most effective antagonist against Diplodia sp [38-41].
Conclusion
The present study revealed that the antagonists were more or less efficient towards each pathogen. Our results were authenticated as in their study it was revealed that different antagonists may be more adapted to the variable conditions on leaves and fruits. Therefore, different concentrations of bacterial antagonists were applied for the protection against Penicillium, Alternaria, Fusarium and Diplodia sp. So, Bacillus and Pseudomonas were proved to be most efficient antagonistic among all isolated epiphytes as was also proved by Yu in his study, that these two epiphytes are efficient bio-agents due to the direct action of chitinase. Also, bacteria show diverse antagonistic mechanism toward phytopathogen fungi, notably space and nutrient competition, hydrolytic enzymes, induction of resistance, volatile compound synthesis, and biofilms.
Acknowledgement
I, Aqleema Banoo, like to acknowledge Division of plant pathology Skuast-K for providing me the essential necessities that were required to conduct this research methodology. I also like to acknowledge the other authors for useful suggestions while preparing the manuscript.
REFERENCES
- Nunes C. Biological control of postharvest diseases of fruit. Eur J Plant Pathol. 2012;133:181-196.
- Brook C, Cooley JS. Temperature relations of apple rot fungi. J Agric Res. 1917;8: 1-103.
- Jan MY. Studies on extend and nature of postharvest spoilage of apple in Kashmir. M. Sc Agriculture (Plant Pathology) Thesis submitted to Sher-e-Kashmir University of Agriculture Science and Technology, Srinagar, Jammu and Kashmir. 1989:1-86.
- Jonsson A, Nybom H, Rumpunen K. Fungal disease and fruit quality in an apple orchard converted from integrated production to organic production. J Sustain Agric. 2010;34:15-37.
- Grahovac M, Indic D, Tanovic B, Hrustic J. Gvozdenac, S. Integrated management of causal agents of postharvest fruit rot of apple. Pesticides andPhytomedicine. 2011;26:289-299.
- Lennox CL, Spotts RA. Sensitivity of populations of Botrytis cinerea from pear-related sources to benzimidazole and dicarboximide fungicides. Plant Dis. 2003;87: 645-649.
- Korsten L, Janisiewicz W. Biological control of postharvest disease of fruits. Annu Rev Phytopathol. 2002;40:411-441.
- Leben C. The bud in relation to the epiphytic microflora. In: Ecology of leaf surface microorganisms[Preece, T. F. and Dickinson, C.H. (Eds)] London Academic Press, UK. 1971;117-127.
- Sharma RR, Singh D, Singh R. Biological control of post-harvest diseases of fruits and vegetables by microbial antagonistic. Biol Control. 2009;50:205-221.
- Gholamnejad J, Etebarian HR, Sahebani N. Biological control of apple blue mold with Candida membranifaciens and Rhodotorula mucilaginosa. Afr J Food Sci. 2010;4:001-007.
- Janisiewich WJ. Post-harvest biological control of blue mold on apples. Phytopathology. 1987;77:481-485.
- Sobiczewski P, Bryk H, Berezynski S. Evaluation of epiphytic bacteria isolated from apple leaves in the control of postharvest diseases. J Fruit Ornam Plant Res. 1996;4:35-45.
- Janisiewicz WJ, Roitman J. Biological control of blue mold and gray mold on apple and pear with Pseudomonas cepacia. Phytopathology. 1988;78:1697-1700.
- Wisniewski M, Wilson C. Biological control of postharvest diseases of fruits and vegetable. Hort Sci., 1992;2:94-98.
- Sobiczewski P, Bryk H. Bacteria for control of postharvest diseases of fruits. In: Environmental biotic factors in integrated plant disease control. Phytopathol. 1995;25:105-111.
- Janisiewicz WJ, Tworkoski TJ, Kurtzman CP. Biocontrol potential of Metchnikowia pulcherrima strains against blue mold of apple. Phytopathology. 2001;91:1098-1108.
- Abano EE, Amoah LK. Application of antagonistic micro-organism for the control of post-harvest decays in fruits and vegetables. Int J Adv Boil Res. 2012;2:1-8.
- Barkai-Golan R. Biological Control. In: Post-harvest diseases of fruit and vegetables: Development and Control. Elsevier, Amsterdam. 2001;147-188.
- Wattal SW. Studies on the nature and extend of postharvest spoilage of apple in Jammu. M. Sc Agriculture thesis submitted to Sher-e-Kashmir University of Agriculture Science and Technology, Srinagar, Jammu and Kashmir. 1993;1-78.
- Aneja KR. Experiments in Microbiology, Plant Pathology and Biotechnology, New Delhi, India, New Age International Publisher. 2004.
- Chowdhry PN, Lal SP, Nati M, Singh DV. Identification of plant pathogenic and biocontrol fungi of agricultural importance. Centre of Advanced Studies in Plant Pathology, Division of Plant Pathology, Indian Agricultural Research Institute, New Delhi. 2000.
- Barnett HL, Hunter BB. Elustrated genera of imperfect fungi. 3rd eddition. Burgess Publishing Co. 1972;1-299.
- Gerhardt P. Manual of Methods for General Bacteriology (Ed. R.G.E. Murray). American Society for Microbiology, Washington, USA. 1981;1-524.
- Huang HC, Hoe JA. Penetration and infection of Sclerotinia sclerotiorum by Coniothyrium minitins. Can J Bot. 1976;54:406-410.
- Fokkema NJ. Fungal antagonism in the phyllosphere. Ann Appl Biol.1978;89:115-117.
- Skidmore AM. Interactions in relation to biological control of plant pathogens. Microbiology of Aerial plant Surfaces, Academic press, London. 1976;507-528.
- Korsten L, Jegar ES, Villers EF. Evaluation of bacterial epiphytes isolated from avpcado leaf and fruit surfaces of biocontrol of avocado postharvest diseases. Plant Dis. 1995;79:1149-1156.
- Schiewe A, Mendgen K. Identification of antagonists for biological control of the post-harvest pathogens Pezicula malicorticis and Nectria galligena on apples. J Phytopathol. 1992;134:229-237.
- Narayanan R, Adorisio D. Modal studies on plate girth. J Strain Anal Eng. 1983;18:111-117.
- Gomez KA, Gomez AA. Statistical Procedure for Agricultural Research 2nd Edition. A Wiley Inter Science Publication, John Wiley and Sons, Inc., New York. 1984;1-680.
- Holt JG, Sneath PHA, Stanley JT, Williams ST. Bergeys Manual of Determinative Bacteriology (9th Edn,). Williams and Wilkins, Baltimore, USA. 1994.
- Ogwu MC, Osawaru ME. Comparative study of microflora population on the phylloplane of okra (Abelmoschus esculentus). Niger J Biotechnol. 2014;28:17-25.
- Bryk H, Dyki B, Sobiczewski P. Inhibitory effect of Pseudomonas sp. On the development of Botrytis cinerea and Penicillium expansum. Plant Prot Sci.2004;40:128-134.
- Grahovac MS, Rahovac JS, Balaz JA, Grahovac JM, Donic BB. Screening of antagonistic activity of selected microorganisms against apple rot pathogens. Rom Biotechnol Lett. 2014;19:8959-8965.
- Gholamnejad J, Etebarian HR, Roustaee AR, Sahebani NA. Biological control of apple blue mold by isolates of Saccharomyces cerevisiae. J Plant Prot. 2009;49:270-275.
- Pusey PL, Wilson CL. Post-harvest biological control of stone fruit brown rot by Bacillus subtilis. Plant Dis. 1984;68:753-756.
- Dukare AS, Sangeeta P, Eyarki NV, Ram KG, Rajbir S. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: A review. Crit Rev Food Sci Nutr. 2018;30: 1498-1513.
- Mendgen K, Schiewe A, Falconi CJ. Biological control of plant diseases. Plant Protect. 1991;45:5-20.
- Yu T, Wang L, Yin Y, Wang Y, Zheng X. Effect of chitin on the antagonistic activity of Cryotococcus laurentii against Penicillium expansum in pear fruit. Int J Food Microbiol. 2008;122:44-48.
- Safdarpour F, Khodakaramian G. Assessment of antagonistic and plant growth promoting activities of tomato endophytic bacteria in challenging with Verticillium dahlia under in-vitro and in-vivo conditions. Biol J Mic. 2019;7:77-90.
- Dukare AS, Sangeeta P, Eyarki NV, Ram KG, Rajbir S. Exploitation of microbial antagonists for the control of postharvest diseases of fruits: A review. Crit Rev Food Sci Nutr. 2019;59(9):1498-1513.
Citation: Banoo A, Shahnaz E, Banday S, Rasool R, Bashir T, Latif R (2020) Studies on Predominant Epiphytic Micro-flora as Antagonists to Post-harvest Pathogens of Apple. J Plant Pathol Microbiol. 11:512. doi: 10.35248/2157-7471.20.11.512
Copyright: © 2020 Banoo A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

