Indexed In
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2024) Volume 8, Issue 2
Stress-Induced Eukaryotic Translational Regulatory Mechanisms
Dilawar Ahmad Mir*, Zhengxin Ma, Jordan Horrocks and Aric RogersReceived: 24-May-2024, Manuscript No. JCMS-24-25940; Editor assigned: 27-May-2024, Pre QC No. JCMS-24-25940; Reviewed: 10-Jun-2024, QC No. JCMS-24-25940; Revised: 17-Jun-2024, Manuscript No. JCMS-24-25940; Published: 24-Jun-2024, DOI: 10.35248/2593-9947.24.8.277
Abstract
The eukaryotic protein synthesis process entails intricate stages governed by diverse mechanisms to tightly regulate translation. Translational regulation during stress is pivotal for maintaining cellular homeostasis, ensuring the accurate expression of essential proteins is important for survival. This selective translational control mechanism is integral to cellular adaptation and resilience under adverse conditions. This review manuscript explores various mechanisms involved in selective translational regulation, focusing on mRNA-specific and global regulatory processes. Key aspects of translational control include translation initiation, which is often a rate-limiting step, and involves the formation of the eIF4F complex and recruitment of mRNA to ribosomes. Regulation of translation initiation factors, such as eIF4E, eIF4E2, and eIF2, through phosphorylation and interactions with binding proteins, modulates translation efficiency under stress conditions. This review also highlights the control of translation initiation through factors like the eIF4F complex and the ternary complex and also underscores the importance of eIF2α phosphorylation in stress granule formation and cellular stress responses. Additionally, the impact of amino acid deprivation, mTOR signaling, and ribosome biogenesis on translation regulation and cellular adaptation to stress is also discussed. Understanding the intricate mechanisms of translational regulation during stress provides insights into cellular adaptation mechanisms and potential therapeutic targets for various diseases, offering valuable avenues for addressing conditions associated with dysregulated protein synthesis.
Keywords
mRNA; Stress; mTOR; Signaling; Translation regulations mechanisms
Introduction
The stress response in cells triggers a cascade of changes in gene regulation, encompassing transcription, mRNA processing, and translation [1-3]. Among these changes, translation stands out as the primary mechanism affected by cellular stress [4]. Adapting translation becomes imperative to confront the challenges imposed by stressful cellular conditions. The regulatory components of the translational machinery play a pivotal role in governing protein synthesis. Maintaining the utmost accuracy in protein synthesis is essential for cellular functions, necessitating the highest level of translational fidelity. The first and most direct mechanism impacted by cellular stress is translation [5]. Translation adaptation is necessary to react to the challenges and obstacles imposed by conditions of cellular stress. In the regulation of protein synthesis, translational machinery components are actively involved. Particularly, initiation emerges as an important target for regulation during stress, given its significant influence on the overall translation rate [6]. Accuracy of protein synthesis is vital for life; consequently, to achieve the requirement of cellular functions, the highest degree of fidelity of translation is mandatory. Various mechanisms produced within cells lead to a global repression of translation under diverse stress conditions, primarily occurring during the initiation process [3,7-9]. Translational machinery components are actively participating in the regulation of proteins synthesis. Even in small microorganisms, the translation initiation mechanism requires more than one hundred macromolecules. Repression of cap recognition and the reprocessing of the ternary complex through eIF2α phosphorylation are critical steps by which cells globally inhibit translation initiation [10]. While significant attention has been paid to translational control at the elongation stage under stress, initiation control, and elongation both rely on the action of the Target of Rapamycin (TOR). Under mTORC1 stress, direct phosphorylation of eIF2α and ribosomal S6 kinase, along with eIF4E dephosphorylation and sequestration by binding proteins (4EBPs), are known to regulate global mRNA translation. However, these regulatory pathways alone do not entirely account for the observed degree of translational repression. The concept of translation reprogramming aligns with the regulation of translation during stress, enabling specific mRNA translation to maintain stress protein expression while halting overall protein synthesis. The regulatory processes can either affect global translation or the translation of specific sequences, and both will be treated independently in the manuscript. The manuscript could be split more obviously into 1) global regulation; and 2) selective regulation. This review provides an overview of how the cellular translational machinery of higher eukaryotic (Human/mammalian systems, yeast, and plants) responds to stress conditions and delves into the forms of translational modification induced by stress. It emphasizes the mechanisms of major signaling pathways involved in translational regulation during various stress types. The focus spans from well-established translational reprogramming in stress responses to recent advancements in understanding initiation and elongation modes of regulatory mechanisms. Additionally, we underscore the impact of translational control on cellular proteostasis, emphasizing processes that could potentially disrupt the aging process.
1. Overview of the Eukaryotic Translation Process
The translation of Messenger RNA (mRNA) constitutes a complex, energy-demanding process characterized by multiple steps and factors [11,12]. It involves the decoding of triplet nucleotide codes on mRNA by tRNA and ribosomes to synthesize polypeptide chains. Translation initiation necessitates coordinated interactions between eukaryotic mRNAs and translation initiation factors, along with the 40S ribosomal subunit [13,14]. The m7GpppN cap structure, present on nuclear-transcribed cytoplasmic eukaryotic mRNAs, plays a critical role in splicing, mRNA stability, polyadenylation, and translation. This cap structure's association with eIF4F is an initial step in mRNA translation, comprising three subunits-eIF4A (a DEAD-box RNA helicase), eIF4E (a cap-binding protein), and eIF4G (a molecular platform with multiple docking sites). The primary function of the eIF4F complex is to recruit ribosomes to mRNA molecules.
mRNA translation involves four stages: Initiation, elongation, termination, and ribosome recycling [15]. Initiation aims to prime the 40S ribosomal subunit for mRNA binding, recruit it to the mRNA, and position it at the initiation codon before the joining of the 40S complex with the 60S ribosomal subunit. Recruitment of the 40S ribosome subunit during translation initiation is deemed essential [14]. This intricate process, involving numerous initiation factors and accessory proteins, is considered the rate-limiting step in mRNA translation [16]. Protein-RNA and protein-protein interactions play pivotal roles in translation initiation [17,18]. Notably, among the twelve distinct factors involved in eukaryotic translation initiation, DEAD-box RNA helicases, eIF4A, and Ded1p (DDX3 in humans) are highly conserved [7,19,20]. The interplay of eIF4A with eIF4E (m7G-cap-binding protein) and eF4G (a scaffolding protein) at the 5′-end of mRNAs is vital [14,19,20]. eIF4G orchestrates initiation by facilitating the recruitment of additional factors, providing a scaffold for ribosome/mRNA bridging [21]. eIF4A helicase unwinds RNA duplexes in vitro and stimulates ATP-dependent RNA unwinding activity in interaction with eIF4G's MIF4G domain [22-24]. The basic translation initiation process is shown in Figure 1.
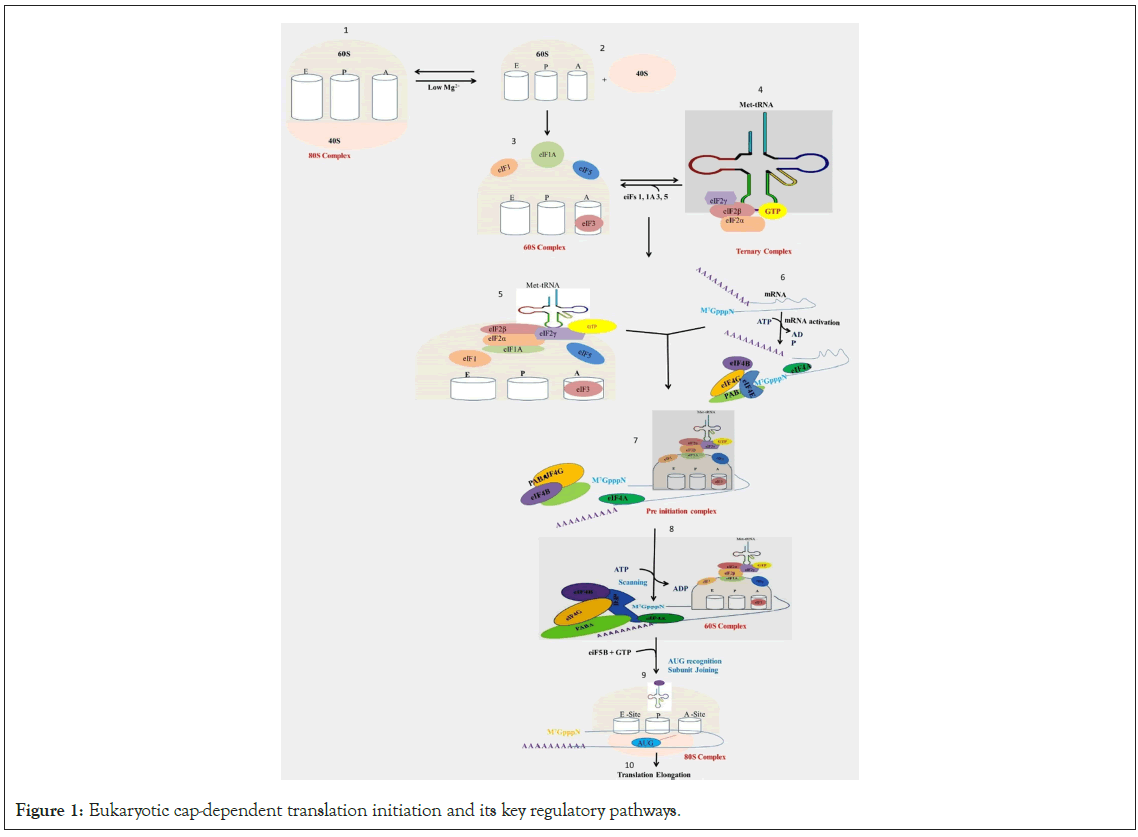
Figure 1: Eukaryotic cap-dependent translation initiation and its key regulatory pathways.
Legend Figure 1: Eukaryotic mRNAs comprise a 5′ m7G cap, which is bound by the eukaryotic initiation factor 4F complex (eIF4E, eIF4G, and eIF4A), and the ternary complex (eIF2-GTP- Met-tRNAi). Translation initiation starts with the assembly of the 43S preinitiation complex (PIC), consisting of the 40S ribosomal subunit, the ternary complex, and the initiation factors (eIF1, eIF1A, eIF3, eIF5). The PIC is recruited to the 5′ cap of the mRNA by the eIF4F complex and eIF4B. Binding of eIF4F to the 5′ cap and PABP to its poly(A) tail activates the mRNA. Successively, the 48S initiation complex is formed, and TC delivers Met-tRNA into the P-site of the ribosome. Before joining of the 60S ribosomal subunit to the PIC, all initiation factors are released from the 40S small ribosomal subunit. Finally, eIF5B unites the 40S and 60S ribosomal subunits to form the 80S initiation complex, and translation elongation begins.
Transcription and translation guarantee the precise transfer and placement of genetic information into folded, functional proteins, ensuring their correct positioning within a cell for optimal functionality. Cellular protein synthesis is intricately regulated to match intracellular demands and external conditions. This process aids in minimizing the overall cost of protein production. Among cellular functions, mRNA translation stands out as a notably energy- intensive process, demanding approximately 75% of the cell’s total energy [25]. Reduced mRNA translation increases cellular resource availability, redirecting resources toward cellular maintenance and repair [26]. Quality control mechanisms to prevent the synthesis of defective proteins, including cotranslational mRNA and protein surveillance at the ribosome [16].
Under stressful conditions, cellular pathways regulating ribosome biogenesis and mRNA translation play critical roles in stress survival. Aberrant proteins synthesized during stressful conditions undergo degradation pathways [27,28]. Failure in stress pathways can lead to the accumulation of misfolded proteins, potentially causing various human diseases such as Alzheimer’s, Huntington’s, and Parkinson’s diseases [29-35]. Depletion or inhibition of translation components can reduce the risk of toxic protein accumulation, potentially extending lifespan [36]. Inhibition of mTOR signaling, a key regulator of translation initiation, has shown increased lifespans in various organisms, indicating its essential role as a longevity regulator [37-39].
2. Translational Reprogramming Under Stress
The regulation of gene expression and translation is critical and serves as a fundamental mechanism influencing various cellular processes, including growth, differentiation, and survival. Precise control of transcription and translation is required to maintain optimal levels of essential proteins for correct protein homeostasis. During adverse stress conditions, cells need rapid and efficient changes in mRNA translation. Stress-responsive protein synthesis targets and reprograms global translation and translation of specific mRNA [16]. While variations in mRNA expression levels play an obvious role in determining the cellular level of proteins, several studies indicate a lack of a consistent relationship between these phenomena. Unique abundant mRNAs may exhibit poor translation, and vice versa [40-42]. Alterations in translational machinery components or their availability can lead to mRNA translation arrest. Additionally, subcellular changes in the localization of messenger ribonucleoproteins (mRNPs) significantly impact translation regulation [43].
Different phases of translation possess specific regulatory targets. The primary targets of translational control are the initiation factors eIF2 and eIF4E. Disruption of these factors influences most of the translatome and results in rapid and robust implications, which are characteristic of the integrated stress response [43]. Phosphorylation of eIF2 globally regulates translation by inhibiting eIF2B recycling and its ability to bind Met-tRNAi, an interaction involved in the translation of nearly all mRNAs [44,45]. Signal transduction pathways, responsive to stresses and physiological stimuli, regulate the functions of eIF2 or eIF4E translation initiation factors. Stress-induced changes in translation initiation are reversible mechanisms that restore translation rates after stress withdrawal [43]. In mammals, eIF2α activation occurs via four different protein kinases, leading to an increase in phosphorylated eIF2α (p-eIF2α) during stress, subsequently inhibiting protein synthesis [46]. To redesign its proteome to change gene expression and renovate the essential signaling pathways to control stress and promote cell survival, cell reduce the rate of global translation [46]. Elevated p-eIF2α levels are expected to increase the translation of specific mRNAs encoding pro-survival and stress-responsive proteins, while potentially inhibiting mRNAs encoding housekeeping genes. ATF4, a key Integrative Stress Response (ISR) gene, preferentially translates under stress. ATF4 mRNA contains upstream ORFs (uORFs) in its 5'-UTR, significantly influencing translation efficiency in response to p‐eIF2α levels [47-49]. Inhibition of translation initiation and polysome disassembly cause the accumulation of untranslated mRNPs in the cytosol [50]. These untranslated mRNPs aggregate with various proteins, subsequently; arrested mRNPs condense into non-membrane-bound subcellular compartments termed cytoplasmic Stress Granules (SGs) [51,52]. Cellular stress, like heat shock and oxidative stress, triggers the formation of SGs through eIF2 phosphorylation SGs exist in a dynamic equilibrium with polysomes [50,53]. Notably, the decrease in SG disassembly perpetuates the translationally arrested state of mRNPs [54].
3. tRNAs Fine-Tune Translation Under Stress
Numerous positions bear conserved modifications within the long 70-90 nucleotides of tRNA, contributing to the structural landscape. Among these, specific alterations within the anticodon loop of tRNAs assume pivotal roles in the reprogramming of translation, especially under stress conditions [55]. The kinetics of codon-anticodon interactions, altering transcript expression and stability, are significantly affected by Wobble base modifications [55,56]. Predominantly, methylation catalyzed by tRNA methyltransferases at the wobble nucleotide bases or ribose sugar represents the most prevalent tRNA modification [57]. Notably, methylation at the wobble base within the tRNA anticodon loop plays a significant role in preserving translational fidelity [58,59].
In response to cellular stress, cells exert control over the methylation and demethylation processes of tRNA bases, consequently fine-tuning translational accuracy [60]. Despite being initially considered degradation remnants, recent research has clear on the role of cleaved tRNA fragments in modulating protein synthesis during cellular stress, proliferation, and differentiation [61,62]. The Ribonuclease angiogenin catalyzes stress-dependent cleavage of tRNAs, specifically targeting the anticodon loop, producing tRNA halves termed tRNA-derived stress-induced RNAs (tiRNAs). Underlying this involved network of regulation is the amino acid control (GAAC) pathway, which monitors and responds to stress-induced nutrient scarcity. Accumulation of noncharged tRNAs during nutrient limitations triggers the activation of the GCN-2 gene [63]. Activated GCN-2, stimulated by binding noncharged tRNAs, orchestrates the response to restriction of nutrients [64]. Furthermore, activated GCN-2 facilitates eIF2α phosphorylation, leading to the inhibition of mRNA translation. Its activation also initiates the expression of genes involved in amino acid biogenesis pathways [63,65]. Studies in yeast and C. elegans have elucidated an interplay between the GCN-2 and TOR pathways. In yeast, inhibition of the TOR pathway activates GCN-2, while in C. elegans, GCN-2 regulates the expression of PHA-4, a downstream transcription factor of LET-363 (the orthologue of TOR) [65,66]. Notably, GCN-2's role resembles that of the TOR pathway, contributing significantly to lifespan regulation; loss of GCN-2 function reduces lifespan during amino acid limitation [65]. Collectively, these findings underscore a close interconnection between the TOR and GCN-2 pathways, emphasizing how nutrient restrictions can trigger a sophisticated system governing mRNA translation and longevity regulation. This basic mechanism of regulation is shown in Figure 2.
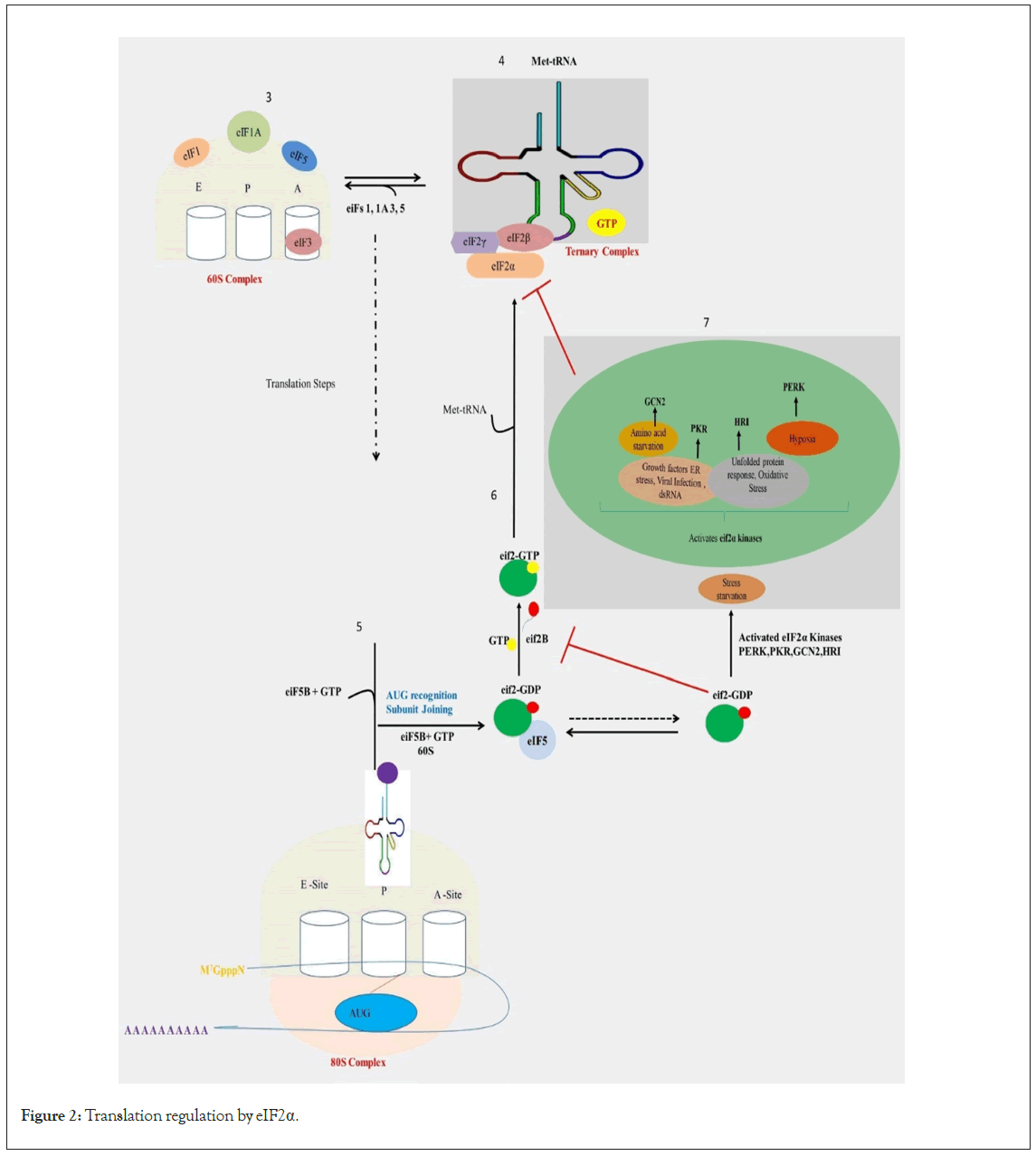
Figure 2: Translation regulation by eIF2α.
Legend Figure 2: Eukaryotic initiation factor 2 alpha is a pivotal factor in the initiation of translation. When eIF2α undergoes phosphorylation, it impedes the assembly of the ternary complex (composed of eIF2, GTP, and initiator tRNA), thereby hindering translation initiation. The eIF2a kinases serve as rapid responders to disruptions in cellular equilibrium. This family comprises four members: PKR-like ER kinase (PERK), double-stranded RNA- dependent Protein Kinase (PKR), Heme-Regulated eIF2a kinase (HRI), and general control nonderepressible 2 (GCN2). Each kinase is triggered by specific environmental or physiological stresses, reflecting their distinct regulatory pathways. PERK, PKR, HRI, and GCN2 kinases are activated by signals such as ER stress, viral infection, and other cellular stressors, leading to the phosphorylation of eIF2 α, a central component of the integrated stress response. Consequently, there is a global attenuation of cap-dependent translation.
4. mRNA Translation Regulation by Chaperones
Hsp27, a key factor in heat shock stress response, plays a vital role in mRNA translation regulation through interactions with eIF4G and PABP1 [67-70]. Under heat shock conditions, eIF4G separates from the PABP1-eIF4G complex and engages with Hsp27, prompting its translocation from the cytosol to the nucleus. This suggests that Hsp27 may facilitate the transfer of Eif4G into the nucleus [68]. Previous research has indicated that following dissociation from PABP1, the eIF4G-Hsp27 complex localizes in insoluble heat shock granules [67]. Recent studies have identified the Hsp27-eIF4G-PABP complex in the cytosol, suggesting that the binding of Hsp27 alone may not solely be responsible for mRNA translation attenuation. At higher temperatures, the interactions between eIF4G and PABP1 with mRNA might be impeded, indicating a decoupling of mRNA nuclear export and translation. This phenomenon might elucidate the observed nuclear accumulation of eIF4G/PABP1 during stress conditions [68]. This data underscores the impact of Heat Shock Proteins (HSPs) on mRNA translation during stress, highlighting their role in reducing new protein synthesis to alleviate the additional burden on chaperones.
5. Translational Regulation of Selective mRNAs during Stress
The process of eukaryotic protein synthesis involves multiple stages, necessitating correspondingly an intricate regulatory system. This multi-stage mechanism is essential to tightly control the translation of critical proteins, as their misexpression could prove lethal to the cell. The association of mRNAs with the translation machinery can either be inhibited or stimulated, constituting a pivotal aspect of translational control. This regulatory landscape encompasses two primary categories; mRNA-specific regulation and global regulation [71]. mRNA-specific regulation selectively influences the translation of particular mRNAs, while global regulation modulates the overall translational efficiency of numerous mRNAs through generalized alterations in the translation process. Despite the documentation of diverse translational regulation processes, only a few are comprehensively understood mechanistically. Translation initiation typically serves as a rate-limiting step, wherein among the translation initiation factors, eIF4E levels are notably low [15].
The formation of the eIF4F complex and subsequent translation initiation represent critical, rate-limiting phases in most circumstances. Extensive research efforts are dedicated to uncovering the molecular underpinnings of this singular regulatory stage. The binding of eIF4E to the 5′ mRNA cap stands as a pivotal step. Additionally, the recruitment of mRNA to the 40S ribosomal subunit is also deemed a rate-limiting initiation step [13,16]. Other significant steps involved in regulating translational initiation encompass the availability of the Ternary Complex (TC) (via eIF2α phosphorylation), modulation of mRNA poly (A) tail length-an enhancer of translation and mRNA stability [72-75]. Furthermore, the control of translation initiation via cap-independent mechanisms, as well as subsequent steps like initiator codon recognition and scanning, may also be subject to modulation [76,77]. Phosphorylation emerges as a key modifier of various initiation factors, often utilized to regulate global rates of protein synthesis. Beyond phosphorylation, a multitude of other posttranslational modifications-such as methylation, glycosylation, and ubiquitination-play vital roles in translational regulation and warrant extensive study [16].
5.1 eIF4F-Mediated 5′-cap regulation
Under normal circumstances, mTOR organizes the assembly of the eIF4F complex at the 5' end of mRNA, facilitating the recruitment of ribosomal subunits and subsequent translation of the transcript [78]. The prevalent mechanism in eukaryotes for regulating translation initiation rates involves the eIF4F method of mRNA 5'-cap recognition. The stability and translational efficiency of eukaryotic mRNAs are notably influenced by the 5' end cap [79-81]. The presence of a methyl group on the 5'-cap is essential for recognition by CBP, eIF4E, and the Decapping Enzyme (DcpS) [82,83]. Decapping serves to deactivate translation initiation and initiates the 5'-to-3' decay of mRNA [84]. Under certain conditions, mRNAs cannot be translated via cap-dependent translation. For instance, cap-dependent translation is inhibited during cellular stress and viral infection [8,85,86]. Approximately 10% of human mRNAs feature 5' UTRs that enable cap-independent translation initiation during stress [15,87-90]. Although the translation initiation factor eIF4F has been traditionally associated with cap-dependent translation, current research investigates stress-specific variations in eIF4F [91-93]. This mechanism of regulation has been illustrated in Figure 3.
5.2 eIF4E regulated ternary complex formation
A ternary complex is a ribonucleoprotein complex containing aminoacylated initiator methionine tRNA, GTP, and initiation factor 2 (eIF2 in eukaryotes, or IF2 in prokaryotes). In prokaryotes, the initiator is fMet-tRNA, while in eukaryotes, it is Met-tRNAi. In eukaryotes, the 43S ribosomal Pre-Initiation Complex (PIC), consisting of the 40S ribosomal subunit and the eIF2-GTP-initiating Met-tRNAi ternary complex, along with additional eIFs such as eIF1, eIF1A, eIF3, and eIF5, is initially recruited to the 5’ terminus of mRNAs. Subsequently, it scans the 5’ Untranslated Region (5’UTR) before moving toward the start codon. Once the start codon is reached, the 60S ribosomal subunit associates with the complex, forming the 80S initiation complex. This complex then facilitates the recruitment of the correct aminoacyl-tRNA into the A (aminoacyl) site, initiating the synthesis of the first peptide bond and transitioning the initiation phase toward elongation illustrated in Figure 1 [94].
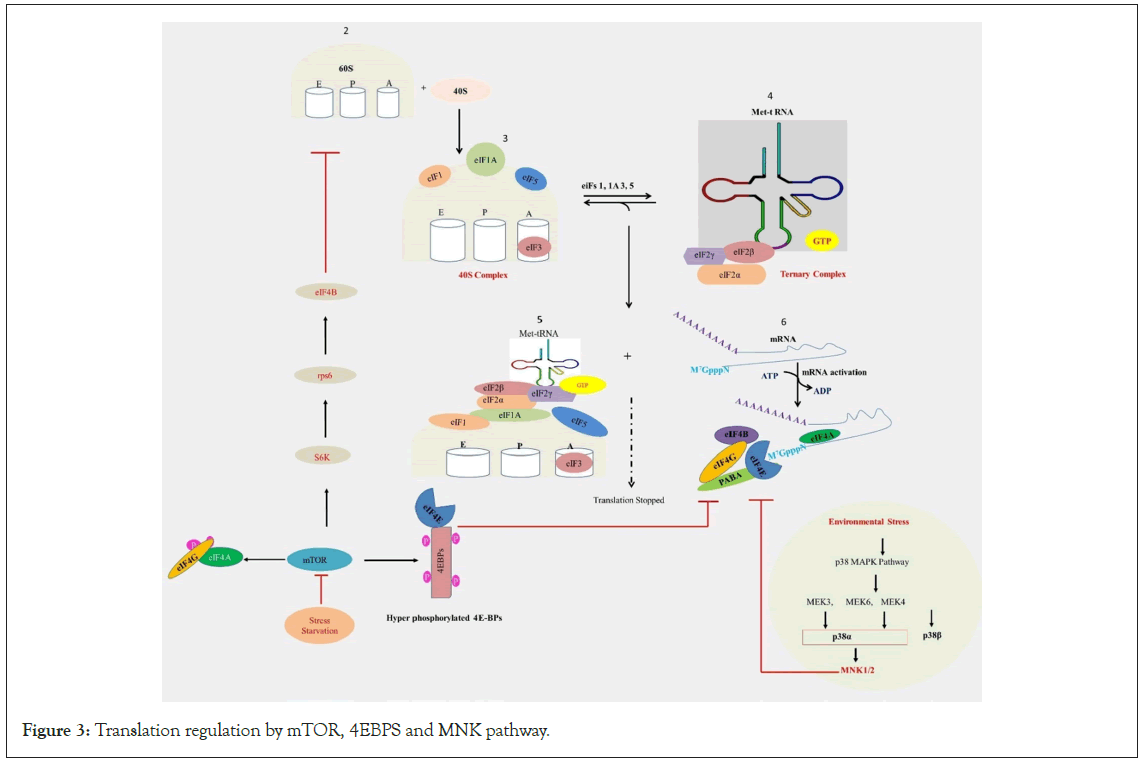
Figure 3: Translation regulation by mTOR, 4EBPS and MNK pathway.
Legend Figure 3: The eIF4F complex plays a vital role in translation initiation, particularly in cap-dependent translation. It comprises three primary subunits: eIF4E: This protein binds to the 5' cap structure of mRNA, serving as the cap-binding protein; eIF4A: An ATP-dependent RNA helicase, eIF4A unwinds the secondary structure of mRNA; eIF4G: Acting as a scaffold protein, eIF4G facilitates interactions between eIF4E, eIF4A, and mRNA. Additionally, there's 4EBP (eIF4E-binding protein), which binds to eIF4E, preventing its interaction with eIF4G and thereby obstructing eIF4F complex formation. Under stress or starvation conditions, 4EBP binding to eIF4E inhibits translation initiation, while eIF2α phosphorylation reduces ternary complex formation. mTORC1 promotes the hyper-phosphorylation of 4EBP, preventing its inhibitory association with eIF4E, thus facilitating eIF4F complex assembly. Furthermore, mTOR enhances the phosphorylation of eIF4G and eIF4B, either directly or via S6 kinases. Given that eIF4E is the most limiting subunit of the eIF4F complex, its availability is essential for recruiting eIF4A to mRNA. eIF4E activity is regulated by MAPK pathways, which directly phosphorylate eIF4E via the MNK protein kinases.
The translation initiation factors eIF4E plays a vital role in facilitating the association of the eIF4F complex with the 5' cap structure of mRNA, serving as a significant rate-limiting factor in the initiation of canonical protein synthesis. Various proteins in animals exert strict control over eIF4E translational activity through phosphorylation or direct binding to eIF4E [95]. Among the well-known regulators of eIF4E translation are 4EBP which bind to distinct lateral and dorsal sites of eIF4E [96-99]. The comprehensive mechanism of 4EBP translation initiation regulation has been illustrated in Figure 3. The interaction between 4EBPs and eIF4E closely correlates with their phosphorylation status, regulated by mTOR [100]. When in a hypophosphorylated state, 4EBP forms a tight complex with eIF4E. The phosphorylation of eIF4E at the Ser-209 residue by the kinases Mnk1 and Mnk2, triggered by tumor promoters, growth factors, and mitogens, significantly influences its oncogenic activity [95,101,102]. The phosphorylation of eIF4E has been shown to selectively regulate the translation of specific mRNAs encoding proliferation, pro-survival (such as BIRC2 and Mcl-1 mRNAs), angiogenesis-related proteins (e.g., VEGFC), and extracellular matrix proteins (MMP3, MMP9) [103]. Studies indicate that eIF4E phosphorylation enhances cellular tolerance to stress, including oxidative and cytotoxic stress, thereby promoting cell survival, regeneration, proliferation, and tumor progression. This process seems to operate through interactions with 4E-Transporter (4E-T) protein and the qualitative control of protein synthesis [104]. It is imperative to conduct detailed research into the interactions between eIF4E phosphorylation, cellular stress, and survival. Furthermore, eIF4E also facilitates the nuclear-cytoplasmic export and degradation of specific mRNAs containing 50-nucleotide elements in their 3' UTR [65,105-107].
Proteins such as LRPPRC, PRH, or 4E-T interact with eIF4E through the canonical eIF4E-binding motif, underscoring the critical role of this domain in eIF4E binding and regulation [101,108,109]. Both eIF4E and 4E-T are components of Stress Granules (SGs) and Processing Bodies (PBs), yet the role of phosphorylated eIF4E in these intracellular structures remains insufficiently studied. Furthermore, other proteins serving as 4E-interacting partners establish connections between eIF4E and 4E-binding proteins via canonical eIF4E-binding motifs or related structures [110]. For instance, Xenopus Maskin and DrosophilaCup act as mRNA-specific 4EBPs, and in the nervous system, Neuroguidin is identified as a 4EBP [111,112]. These specific eIF4E interacting partners play vital roles in regulating animal development programs, primarily by facilitating various protein-protein interactions that link the 5' and 3' UTRs of specific mRNAs, rendering them translationally inactive [113,114].
5.3 eiF2 and regulation of translation initiation
Accurate translation initiation is a vital process for cells to synthesize the correct proteins [7,78]. The eIF2 protein plays a pivotal role in protein synthesis by binding to initiator tRNA (tRNAi Met) within the cytoplasm and transporting it to ribosomes, where it recognizes the AUG (Adenine-Uracil-Guanine) codon on mRNA. The TC complex (eIF2–GTP–tRNAi Met) serves as a critical intermediary in the translation initiation pathway. Comprising three subunits (α, β, and γ), eIF2 is a heterotrimeric protein. The eIF2γ core subunit interacts with GDP/GTP and tRNAi Met, while eIF2α facilitates AUG codon recognition and tRNAi Met-binding to the subunit. EIF2β interacts with eIF2 ligands and other factors essential for the regulation of eIF2 function [115-118].
Extensive research has explored the inhibition of protein synthesis through eIF2α phosphorylation. During stress, phosphorylation of the eIF2α subunit impedes eIF2β's ability to exchange Guanosine Diphosphate (GDP) for Guanosine Triphosphate (GTP). Consequently, the formation of active TC is significantly reduced, leading to global translation downregulation [4,71]. Phosphorylated eIF2α-GDP competitively inhibits eIF2β, as it exhibits a higher affinity for eIF2β compared to unphosphorylated eIF2α-GDP, elucidating the molecular mechanism behind this inhibition [119]. Four types of kinases phosphorylate the conserved Ser51 residue of the eIF2α subunit in response to diverse stresses [115,120]. In eukaryotes, phosphorylation of the eIF2α subunit during stress constitutes a primary mechanism for regulating mRNA translation. In yeast, GCN2 kinase phosphorylates the eIF2α subunit under nutrient starvation conditions [63,121]. Similarly, during nutrient deprivation, protein misfolding, or immune responses in mammals, GCN2 phosphorylates eIF2α [122-124]. The global translational response to stresses involves a critical mechanism of eIF2-GTP regeneration [125]. Recently, a fail-safe regulatory switch has been identified, wherein eIF2β binds to and inhibits phosphorylated TC and TC/eIF5 complexes, offering an alternative pathway to inactivate eIF2/eIF2β complexes [10]. This mechanism of translation initiation regulation has been illustrated in Figure 2.
6. Translation Elongation Regulation and Signaling Pathways
As discussed earlier, the initiation phase has been considered the predominant target for translational regulatory mechanisms. However, accumulating evidence suggests that control can also be exerted at the elongation and termination phases [126]. One common regulatory mechanism involves the phosphorylation of elongation factors, like some initiation factors. Phosphorylation of the elongation factor eEF2 on the Thr56 amino acid residue within the GTP-binding domain is a well-known regulatory mechanism in response to oxidative stress. This phosphorylation alters its affinity for the ribosome, leading to its inactivation [127-130]. Various stress signals activate eEF2 kinase (eEF2K) by AMPK-mediated phosphorylation on the serine 398 residue. eEF2K is a member of the α-kinase family, and its activation or inactivation is regulated by anabolic and mitogenic signaling pathways, such as mTORC1 and the Mitogen-Activated Protein (MAP) kinase pathway [131,132]. To facilitate the rapid continuation of translation elongation, eEF2K is degraded by the ubiquitin-proteasome system. The regulation of translation elongation by mTOR signaling at eEF2 has been extensively reviewed [133,134]. Additionally, eEF2 activity can be modulated by Cytoplasmic Polyadenylation Element Binding protein 2 (CPEB2), an RNA-binding protein that reduces the GTP hydrolysis of eEF2 [135]. The mechanism of translation elongation regulation has been illustrated in Figure 4.
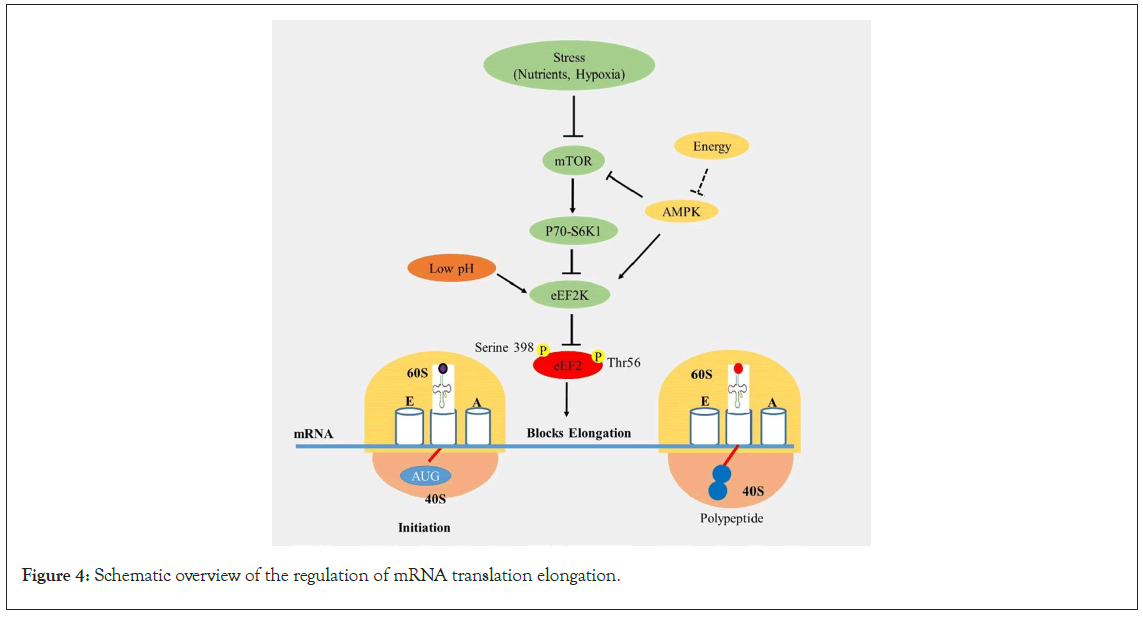
Figure 4: Schematic overview of the regulation of mRNA translation elongation.
Legend Figure 4: Upon 80S complex formation, the ribosome is primed for translation elongation. eEF2 is regulated by the mTORC1 and MAP kinase signaling pathways by controlling eEF2K during stress. eEF2K is activated or inactivated by phosphorylation at Thr56 and Serine 398 residues by anabolic and mitogenic signaling agents (mTORC1 and MAP kinase pathway). Acidic conditions inhibit the phosphorylation reaction catalyzed by eEF2 kinase (eEF2K) and block translation.
Under normal conditions, CPEB2, through binding to the 3′UTR, reduces the translation of HIF1α mRNA. Interestingly, CPEB2 dissociates from HIF-1α mRNA under hypoxic conditions, allowing for the synthesis of HIF-1α and adaptation to hypoxia. Furthermore, eEF2 activation is suppressed during amino acid starvation stresses, endoplasmic reticulum stress, energy stress caused by hypoxia, and genotoxic stress [130,136-138]. Uncharged tRNA accumulation can stall elongating ribosomes in these conditions and play a role in activating eIF2 kinase Gcn2 [139]. Recently, the binding of modified or damaged tRNA has been revealed during stress, contributing to both processes [140].
7. Developing Concepts in Translational Regulation
Considerable attention has been directed towards the regulation of mRNA post-transcriptional modifications for over a decade. The stability and translation of target mRNAs are also influenced by the more recently identified processing bodies and small RNAs cytoplasmic processing bodies and small RNAs. Herein, we expound upon these novel concepts, emphasizing their role in translational regulation. Furthermore, we examine recent examples illustrating how the modulation of alternative transcripts can impact translation.
7.1 P-bodies and translation regulation mechanisms
Processing bodies (P-bodies) and Stress Granules (SGs) are cytoplasmic Ribo-Nucleo-Protein (RNP) granules that primarily constitute pools of translationally repressed mRNAs and proteins associated with mRNA decay, thereby indicating their roles in transcriptional regulation [141,142]. P-body mRNPs consist of translation repression factors i.e. GW182, Pat1, Dhh1/RCK/ p54, Caf1/Pop2, Edc3, Xrn1 and eIF4E and eIF4G, while SG mRNPs encompass a subgroup of translation initiation factors, potentially poised for re-entry into translation [143]. Notably, stress and genetic variations dictate the composition of P-bodies, SGs, and related RNA granules. These cytoplasmic granules are highly dynamic, with their formation and disassembly [144,145]. They are largely conserved in all eukaryotic cells and exhibit characteristics of liquid droplets. However, the precise role of these granules in translational repression or mRNA decay remains inadequately resolved. The specific mechanism governing the transport of these mRNAs into P-bodies for translational repression remains unidentified. Phase transition techniques have been employed to investigate how soluble RNPs condense into liquid or solid bodies [146-148]. These RNP bodies functionally regulate RNP exchanges, subcellular localization, and concentration [149,150]. Various pieces of evidence indicate a connection between P-bodies and translation. Under environmental conditions, mRNAs exist in two distinct states: Associated with polysomes, actively translated, or associated with P-bodies, translationally repressed P-bodies' size and number vary depending on cellular conditions, such as glucose depletion, osmotic stress, UV light, and acid stress [151,152].
Amino acid deprivation has been demonstrated to induce the formation of P-bodies in mammalian cells, along with the localization of mRNA within these structures. This evidence strongly supports the idea that the translation rate has a direct impact on the number of P-bodies. Initially, P-bodies were considered sites for mRNA decay, primarily due to their association with decapping complexes [153]. P-bodies are increasingly recognized as temporary repositories for translationally silenced mRNAs that, without undergoing decay, can re-enter the translating pool particularly in processes like early development, oogenesis, and neuron plasticity [154-157]. Several studies suggest that translation repressors are essential, but decay machinery is not, for P-body formation [158]. P-bodies can be disrupted without halting decay [159,160]. Although P-bodies might serve as centers for controlling mRNA destiny, the identity of P-body mRNAs and their specific role remain poorly defined [161].
7.2 Stress granules and translational regulation mechanisms
Cytoplasmic Stress Granules (SGs) complex combining with mRNA translational regulatory pathways during periods of stress [162]. SGs are delineated as stress-induced membraneless, phase-dense cytoplasmic bio-condensates comprising translationally silent mRNAs associated with preinitiation complexes i.e. RNA translation initiation factors (eIFs), and RNA-Binding Proteins (RBPs), precipitating assembly when translation initiation is obstructed [163]. Notably, SGs lack eIF5 and eIF2, essential proteins for the transformation of ribosomal preinitiation complexes into translationally competent ribosomes [164]. Following the resolution of stress, SGs can transition back into polysomes [165-167]. SGs are believed to form through liquid-liquid phase separation, driven by cooperative interactions among RNA-RNA, protein-RNA, and protein-protein interactions [168-170]. Despite analogies to P-bodies and some common components, SGs contain specific constituents, including translation initiation factors, 40S ribosomal subunits, and Antioxidant Response Element (ARE) binding proteins [54]. However, fusion events and close associations between SGs and P-bodies are evident [54,171]. Formation of stress granules has been illustrated in Figure 5.
Stress-induced eIF2α phosphorylation is both essential and sufficient for SG assembly [53,167]. As discussed earlier, stress-induced phosphorylated eIF2α inhibits eIF2B activity, halting translation initiation and promoting SG assembly [9,45]. Under stress, various transcripts that escape eIF2α phosphorylation-mediated translational arrest produce proteins protecting cells from stress-induced damage. For instance, HSP70 mRNAs are selectively translated and excluded from SGs [167,172]. The Integrated Stress Response (ISR) kinase family, associated with eIF2α phosphorylation during stress, senses charged tRNAs through GCN2, dsRNA through PKR, ER stress through Protein Kinase-Like ER Kinase (PERK), and redox state and heme stress through HRI [124,173-175]. Deletion or inactivation of these eIF2α kinases renders cells insensitive to the respective stresses, with each kinase triggered by distinct stress types, subsequently leading to SG formation [123]. Following SG assembly, pharmacological manipulations, such as the PERK signaling inhibitor ISRIB (Integrated Stress Response Inhibitor), can reverse eIF2α phosphorylation by activating eIF2B. ISRIB treatment has been shown to reverse SG assembly and readily restore translation [176,177].
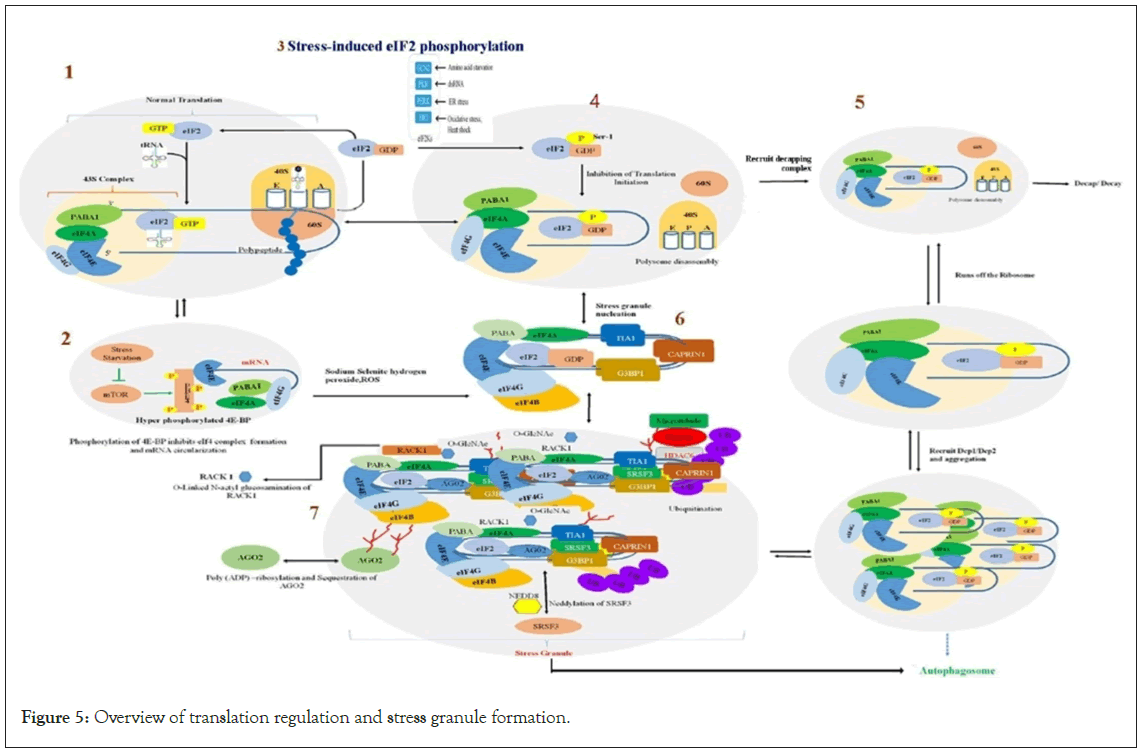
Figure 5: Overview of translation regulation and stress granule formation.
Legend Figure 5: Transcribed RNAs form nuclear messenger ribonucleoprotein particles (mRNPs). RNA-binding proteins linked with mRNA are shifted to the cytosol, where they govern cytoplasmic localization and translational proficiency of the mRNA. mRNA conversion into proteins activates the assembly of translation complexes, and when this process is stalled, the mRNPs accumulate as stress granules.
Step 1: In normal translation, the eIF4F complex recruits the 43S ribosomal subunit. Upon recognition of the initiation codon by the anticodon of tRNAMet, eIF2-GTP is hydrolyzed, and eIF2-GDP is released, and early initiation factors are displaced by the 60S ribosomal subunit.
Step 2: Under stress, mTORC1 promotes the hyper-phosphorylation of 4EBP, inhibiting the association of eIF4E with eIF4G and eIF4A to form the eIF4F complex properly, and blocking translation.
Step 3: In stressed cells, phosphorylation of eIF2α by GCN2, HRI, dsRNA, PERK, and/or PKR converts eIF2 into a competitive antagonist of eIF2B, depleting the stores of eIF2/GTP/tRNAMet. This stops the exchange of GDP-GTP and the restoration of the 43S pre-initiation complex, inhibiting translation.
Step 4: Upon eIF2α subunit blockage, elongating ribosomes ‘run-off’ the mRNA.
Step 5: Following translation, mRNAs can quit translation and assemble a translationally repressed mRNP that can be either degraded or assembled into P bodies. mRNAs enclosed by P bodies can be subject to decapping and 5′-3′ degradation, or they can exchange P-body components for stress granule components to revert translation. Before re-entering translation, mRNAs should obtain extra translational components (eIF2, eIF3, and 40S subunits). Specific factors like mRNA binding proteins or the presence of a poly (A) tail might affect this process. So, mRNPs within stress granules can return to translation initiation again and enter polysomes or can be targeted for autophagy.
Step 6: Translation inhibition, which causes the formation and aggregation of stress granules, is a consequence of the marked accumulation of untranslated mRNPs as a result of blocked initiation of translation, and the formation of stress granules themselves is not required for translation arrest. GTPase-activating protein-binding protein 1 (G3BP1) and T cell-restricted Intracellular Antigen 1 (TIA1) attach to the polysome-free mRNAs and build up to nucleate stress granule formation. Sodium selenite or hydrogen peroxide treatment (ROS generation) inhibits the functions of mTOR, resulting in stress granule formation. For granulation, G3BP1 must be dephosphorylated and demethylated. Poly (ADP)- ribosylated stimulates stress granule nucleation. Binding G3BP1 to cell cycle-associated protein 1 (CAPRIN1) encourages the formation of stress granules.
Step 7: Aggregate large stress granules from smaller focal points. This approach includes retrograde microtubule-dependent trafficking mediated by dynein motors and histone deacetylase 6 (HDAC6) binding to G3BP1, microtubules, and polyubiquitin chains enriched in stress granules. Post-translational modifications of stress such as poly (ADP)-ribosylation and O-linked N-acetylglucosamination (O-GlcNAc) control the recruitment of different proteins. The O-GlcNAc-dependent recruitment of a receptor for activated RACK1 protein to stress granules causes RACK1-mediated pro-apoptotic signaling to be sequestered and inhibited. Neddylation, a post-translational modification, is necessary for the formation of stress granules by mediating the Serine/arginine-Rich Splicing Factor 3 (SRSF3) interactions with the eIF4F complex, N-acetylglucosamine, mRNP, messenger ribonucleoprotein.
7.3 miRNAs and translational regulation mechanism
In various biological pathways, two types of small RNAs, microRNAs (miRNAs), and short interfering RNAs (siRNAs) have emerged as significant gene regulators. These non-coding regulatory RNAs promote the stability and translation of mRNA, resulting in repressed gene expression [178]. Both miRNAs and siRNAs are approximately 21-26 nucleotides in length, and their differentiation is based on their biogenesis [179-183]. These miRNAs are derived from precursors of more than 70 nucleotides, which are hairpin segments with poor base-pairing.
In contrast, siRNAs originate from RNA precursors that are perfectly complementary, either post-transcriptionally or co-transcriptionally [182]. The biogenesis of siRNAs is categorized into canonical and non-canonical pathways [183]. Most miRNAs in animals are processed by the consecutive action of the RNase III-like enzymes Drosha and Dicer from longer hairpin transcripts, whereas in plants, only Dicer is involved in this process. The Argonaute family protein (AGO) is loaded onto one strand of the hairpin duplex (forming the core of silencing complexes (miRISCs) induced by miRNA [183]. This complex can silence the expression of target genes at the post-transcriptional level. Bioinformatics studies suggest that the human genome encodes approximately 1000 miRNAs, each regulating about 10 mRNAs, potentially affecting over 30% of all genes [184,185]. miRNAs are known to interact with both the 5' and 3' untranslated regions of target mRNAs, inducing mRNA degradation and translational repression [182,186,187]. Recent studies indicate that miRNAs localize to multiple subcellular compartments to regulate translation and transcription rates [188]. Remarkably, all subcellular sections involved in miRNA-mediated mRNA expression repression seem to concentrate in P-bodies. P-bodies are believed to be sites where mRNAs are stored and occasionally degraded away from the translation machinery [189]. P-bodies contain 5′ and 3′ exonucleases, decapping enzymes, and Ago proteins (GW182 and Rck/p54), suggesting that P-bodies play a role in miRNA-mediated repression or degrade mRNA machinery [141,190]. Cytoplasmic stress granules also contain Ago proteins, miRNAs, and their target mRNAs [191]. The localization of Ago proteins in stress granules and P-bodies is miRNA-dependent, suggesting a potential collaboration between stress granules and P-bodies in miRNA-mediated translation regulation [190]. The mechanism used by miRNAs to regulate target gene expression has been a controversial subject. In vivo and in vitro studies confirm that miRNAs, depending on various factors, can inhibit translation, destabilize mRNA, or both. In both animals and plants, miRNAs can silence targets through RNA degradation as well as translational repression pathways [192]. In animals, binding of miRNA-Induced Silencing Complexes (miRISCs), containing GW182 proteins, to 3′UTR target sequences recruits deadenylation factors, eliminating the poly (A) tail and making the mRNA susceptible to exonucleolytic degradation [193-196].
In plants, a common mechanism involves the perfect pairing of miRNA with its target site, supporting endonucleolytic cleavage of mRNA by Argonaute [197-199]. Both in animals and plants, there are instances where miRNAs causes reduced protein (but not mRNA) levels, suggesting that translational repression is directed by miRISC. Recent studies indicate that the Carbon Catabolite Repression-Negative On TATA-less (CCR4-NOT complex), recruited by GW182, deadenylates target mRNAs, resulting in the repression of translation initiation [200-204]. The exact mechanism causing the inhibition of protein production is not clear, but in animals, it has been proposed to occur at initiation, elongation, co-translational protein degradation, and premature termination of translation [159,185,203,204]. There is also increasing evidence suggesting that miRNAs interfere with the functions of the IF4F complex and PABPC during translation and/or mRNA stabilization [73]. From the discovery of miRNAs, significant progress has been made in understanding how cells produce miRNAs, their regulatory effects on the central dogma, and their involvement in various physiological and pathological events. Recent studies have highlighted that miRNAs can serve not only as biomarkers for diseases but also play a significant role in intercellular communication [205-209]. However, our knowledge regarding when and how miRNAs exert regulatory effects on transcription is still insufficient. Additionally, conditions under which miRNAs cause translational activation need further exploration.
8. Translation Regulation by mTOR through 4EBP and S6K1
Protein translation regulation primarily occurs during two pivotal stages of translation initiation: The recruitment of the 40S ribosomal subunit and the loading of initiator methionyl-tRNA onto the 40S ribosomal subunit [71,210]. The translation initiation factor eIF4E oversees 40S subunit recruitment and is inhibited by the binding proteins 4EBP [111,211]. The 4EBP is an important regulator of overall translation levels in cells. Meanwhile, the eIF2α protein governs tRNA loading, controlling the recycling of eIF2 [71]. mTOR, a member of the phosphatidylinositol kinase family, forms two distinct complexes, mTORC1 and mTORC2 [212]. Reduced mTOR signaling under stress inhibits processes such as Ribosomal protein (r-protein synthesis), Ribosomal DNA (rDNA) transcription, and mRNA translation initiation [213-214]. mTORC1 primarily promotes protein synthesis by phosphorylating two key effectors, eIF4E Binding Protein (4EBP) and p70S6 Kinase 1 (S6K1). This phosphorylation prevents the assembly of the eIF4F complex by 4EBP, which directly interacts with eIF4E [44,215,216]. Under optimal conditions, mTORC1 constitutively phosphorylates 4EBP at multiple sites and their phosphorylated variants (p-4EBPs) cannot bind eIF4E. mTORC1 phosphorylation trigger its dissociation from eIF4E [194,122]. During stress, mTORC1 inactivation leads to dephosphorylation of p-4EBPs. These dephosphorylated 4EBP variants bind to eIF4E, inhibiting eIF4F complex assembly and subsequently leading to translation inhibition. The binding sites of 4EBPs and eIF4G on eIF4E overlap. Another class of proteins governing translation during stress is S6 kinases (S6Ks) [217]. Under optimal conditions, mTORC1 directly phosphorylates S6K1, enabling its activation by PDK1 [218]. Phosphorylated S6K1 targets Ribosomal Protein S6 (RPS6), a component of eIF4B and the 40S ribosomal subunit, which promotes eIF4A's helicase activity [217,219,220]. The role of phosphorylated S6K in facilitating translation remains unclear, although its inactivation by mTORC1 during stress is predicted to inhibit translation [221]. S6K1 phosphorylates and activates various substrates, including eIF4B, a positive regulator of the eIF4F 5' cap binding complex [218,222]. Moreover, S6K1 degrades the eIF4B inhibitor PDCD4 through phosphorylation [223,224]. Interestingly, the binding of mTORC1 with eIF3 promotes interactions between the 40S subunit and eIF4G, enhancing the recruitment of 40S ribosomal subunits to the eIF4F cap-bound complex and facilitating Protein Initiation Complex (PIC) assembly [225]. This orchestrated network involving mTOR and its associated complexes, as well as various kinases and regulatory proteins, intricately governs translation initiation under different physiological conditions. The comprehensive mechanism has been illustrated in a Figure 6.
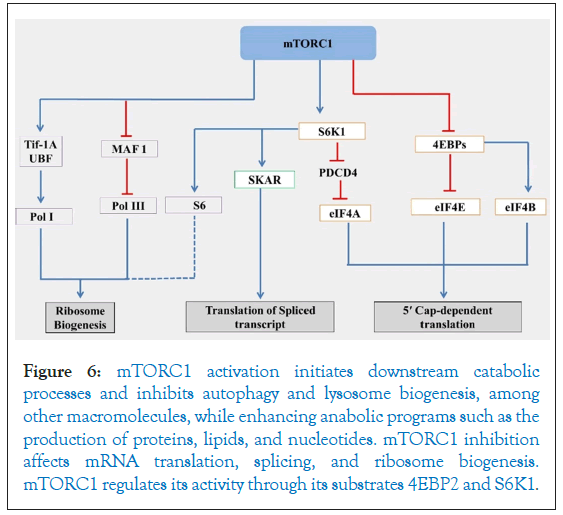
Figure 6: mTORC1 activation initiates downstream catabolic processes and inhibits autophagy and lysosome biogenesis, among other macromolecules, while enhancing anabolic programs such as the production of proteins, lipids, and nucleotides. mTORC1 inhibition affects mRNA translation, splicing, and ribosome biogenesis. mTORC1 regulates its activity through its substrates 4EBP2 and S6K1.
9. Ribosome Biogenesis and Translation Regulation Mechanisms
mTORC1 modulates RNA polymerase III-dependent transcription by interacting with transcription factor-IIIC and Maf1 [226]. Studies have revealed that mTOR directly interacts with ribosomal DNA promoters, leading to chromatin remodeling and subsequent activation of ribosomal gene transcription [227,228]. The growth-dependent Transcription Initiation Factor (TIF-IA) promotes the activation of ribosomal DNA gene transcription by interacting with RNA polymerase I [229]. Inhibition of mTORC1 by rapamycin/ stress leads to TIF-IA inactivation, resulting in the downregulation of 47S pre-rRNA transcription [214]. This comprehensive mechanism has been illustrated in Figure 6. Phosphorylation of eIF2α leads to the formation of Stress Granules (SGs), preferentially inhibiting the translation of mRNAs encoding ribosomal proteins [230]. The mTOR stress-response pathway primarily targets the translation inhibition of ribosomal protein mRNAs and mTOR inactivation further impedes the translation of these mRNAs [212]. Moreover, active mTOR aids in the transcription of RNA Pol I-dependent rRNA genes [231]. Thus, modifications in mTOR activity directly regulate both rRNA transcription and ribosomal protein mRNA translation. Ribosome biogenesis encompasses various steps occurring within three distinct sub-nucleolar components, starting from the initiation of Pol I transcription to the processing of pre-rRNA and eventual ribosomal assembly. Any disruption in this highly orchestrated process can lead to nucleolar stress, resulting in cellular insults [232].
Ribosome biogenesis is a highly energy-consuming process, with studies suggesting that approximately 75% of transcriptional activity is dedicated to this fundamental cellular function [233]. In response to stress, repression of ribosomal protein synthesis has been observed [234]. The orchestration of ribosome biogenesis is intricately regulated by both internal and external signaling mechanisms within the cytoplasm and mitochondria. The generation of ribosomes necessitates the involvement of all three RNA polymerases (I, II, and III). Within nucleoli, RNA polymerase I drives the transcription of the polycistronic 47S pre-rRNA, which upon splicing produces three distinct types of rRNAs (18S, 5.8S, and 28S). Meanwhile, RNA polymerase II and III act in the transcription of genes encoding ribosomal proteins and 5S rRNA, and tRNAs, respectively. Notably, RNA polymerase I-dependent transcription represents the rate-limiting step in ribosome production [235].
Key regulators of ribosome biogenesis are the mTORC1 complex and the c-myc proto-oncogene. Both entities facilitate the expression of 47S pre-RNA by activating transcription factors for ribosomal DNA (selective factor 1) and nucleolar transcription factor (upstream binding), thereby stabilizing the initiation complex through promoter binding [214,231]. Additionally, c-myc stimulates RNA polymerase III activity via transcription factor-IIIB [231]. At promoter regions, c-myc induces histone acetylation (H3 and H4), leading to chromatin decondensation and facilitating rRNA gene transcription [236,237]. Furthermore, c-myc aids in the activation of both small and large ribosomal subunit proteins. The expression of c-myc is dependent on the phosphorylation/dephosphorylation dynamics of the Wnt/GSK-3β/β-catenin signaling pathway, which may impact the efficiency of ribosome biogenesis, although further experimental validation is required [226].
10. Regulation of Translation by Amino Acid Deprivation and mTOR
During Dietary Restriction (DR), protein synthesis is decreased, likely due to reduced activation of the mechanistic Target of Rapamycin (mTOR) kinase [238,239]. A life-long reduction in protein translation, however, slows down aging, prolongs lifespan, and ameliorates cellular senescence and several age-related diseases [240-244]. The crucial response of mRNA translation to stress and longevity is established by observing that knockdown of components of the mTOR pathway or translation initiation factors directly increases longevity in various species, as does reduced S6K activity [245-249]. For example, in S. cerevisiae, inhibition or deletion of ribosomal protein expression increases replicative lifespan [250]. In Drosophila, the d4EBP-1 protein is vital for longevity extension under Dietary Restriction (DR), and its transcription regulation is under the control of the FOXO transcription factor [239,251]. FOXO transcription factor is a conserved longevity regulator downstream of insulin signaling. Insulin signaling can control TOR activity through phosphorylation of S6K and altering the activity of the upstream TSC1/TSC2 regulatory complex. Reduced Insulin/IGF-1 signaling in long-lived Ames dwarf mice and Snell mice leads to reduced protein synthesis [252,253]. Furthermore, in C. elegans, deletion or inhibition of several genes of translation regulators, mainly worm homologs of eIF4E (ife-2), eIF4G (ifg-1), eIF2B (iftb-1), and a number of ribosomal proteins, translation initiation factors increase lifespan [245,254]. Inhibition of C. elegans, ifg-1, results in selective expression of stress response genes and is also down-regulated upon starvation and reduced in long-lived dauers [240,255]. DR is also capable of activating proteins that respond to stress, which may also lead to an improvement in lifespan. Ribosomal protein and S6K knockdowns are independent of DAF-16, while the translation initiation factor may be dependent on DAF-16 to improve longevity, suggesting a complex relationship between the insulin signaling pathway and translation regulation.
To detect and respond to nutrient variations, all species continuously track their immediate environment, and a wide variety of adaptive mechanisms have evolved. How cells respond to nutritional availability and deficiencies by altering genomic information flow, transcriptional and translational levels, which are linked to global protein synthesis inhibition [256,257]. Deprivation of certain nutrients may impose more stringent restrictions on mRNA translation than others to support a particular metabolic function [258]. Amino acids play a prominent role among the key nutrients in controlling the mTORC1 pathway. The amino acids, leucine, and arginine in particular, have been reported to be completely necessary for mTORC1 activation in mammalian cells [259]. Glucose, glycine, glutamine, leucine and serine have been documented to be differently dependent on cells during division [164,260-264]. A restriction of amino acids, however, can also contribute to the activation of stress response pathways and to an improvement in model organism longevity [263,265]. In rodents, it is well known that a diet containing reduced small quantities of methionine substantially increases 45% of the safe lifespan relative to control rats [266,267]. The comprehensive mechanism related to the regulation of translation by mTOR and amino acid deprivation has been illustrated in Figure 7.
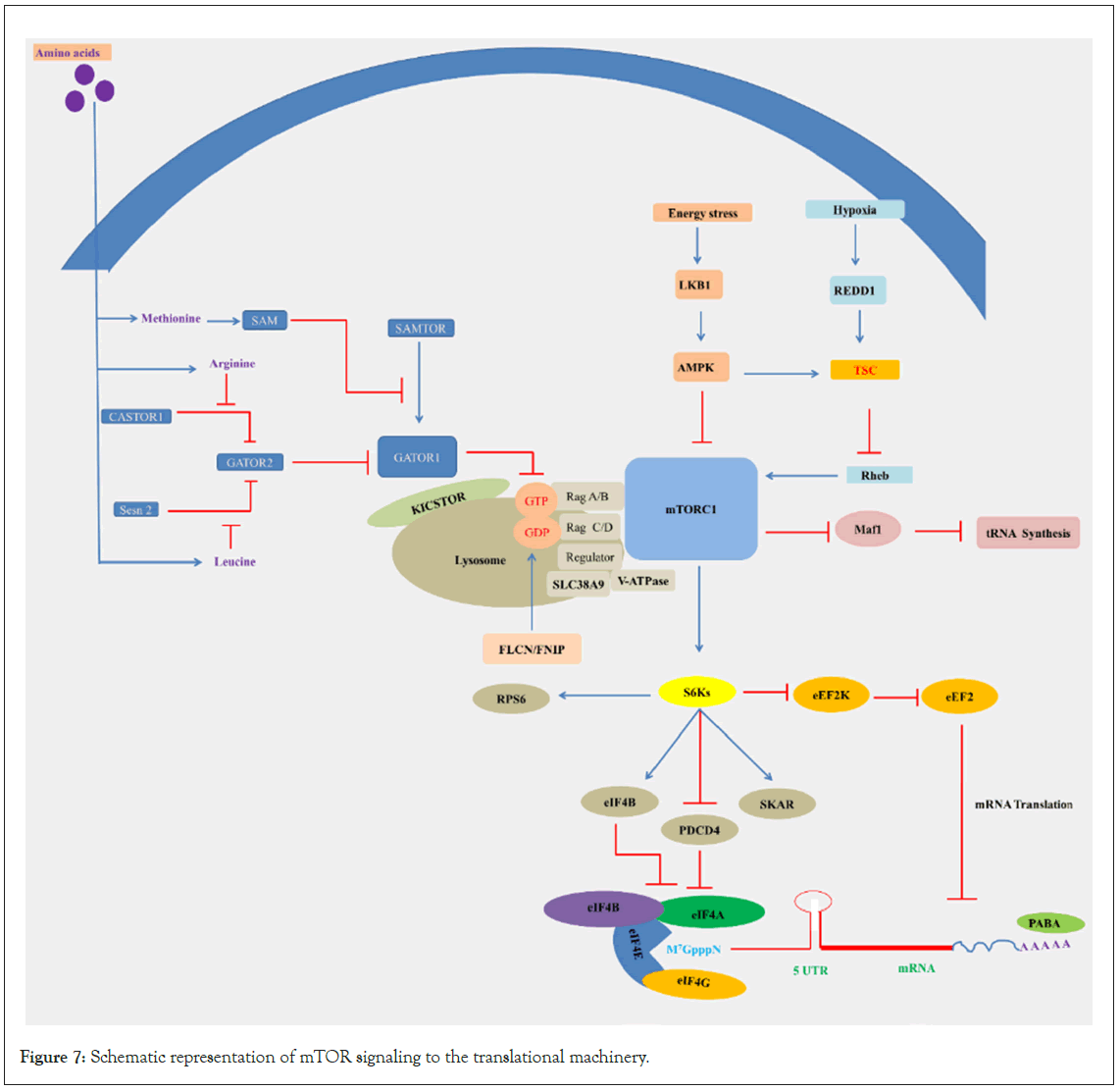
Figure 7: Schematic representation of mTOR signaling to the translational machinery.
Legend Figure 7: Rag GTPases, activated by amino acids, recruit mTORC1 to the surface of the lysosome, and the small GTPase Rheb activates mTORC1 in its GTP-bound state. The availability of amino acids controls the nucleotide state of the Rags, this process depends on the interplay between Ragulator and GATOR1. Ragulator serves as a lysosomal scaffold for RagA/B, and GATOR1 acts as a GTPase-Activating Protein (GAP) for RagA/B. GATOR1 is a critical negative regulator of the mTORC1 pathway. The GATOR2 complex acts in parallel to GATOR1 and is a key positive regulator of the mTORC1 pathway. The amino acid sensors Sestrin2 and SLC38A9 sense cytosolic leucine and putative lysosomal arginine, respectively, for the mTORC1 pathway. SAMTOR and CASTOR2 sense methionine and arginine, respectively, and starvation of these amino acids inhibits the mTORC1 pathway. In the absence of leucine, Sestrin2 interacts with GATOR2 and inhibits mTORC1 signaling, while SLC38A9 forms a supercomplex with Ragulator and is necessary for transmitting arginine, but not leucine, sufficiency to mTORC1. Following activation of the Ras/ ERK pathway, S6K phosphorylates rpS6, eIF4B, PDCD4, and eEF2K, which are important regulators of translation. Low oxygen and energy conditions also diminish protein synthesis. Hypoxia requires the Tuberous Sclerosis Complex (TSC) to downregulate S6K activity. In addition, translation initiation is inhibited under hypoxic conditions by the eIF2α-phosphorylating kinase PERK. Low cellular energy levels activate AMPK, which inhibits mTOR by stimulating TSC2 function.
mTORC1 senses nutrient levels through a sophisticated system [268,269]. When cellular amino acid levels are ample, mTORC1 is activated. The discovery of the Rag GTPases has solved the mystery of how amino acids communicate their availability to mTORC1, as they are an essential component of the nutrient sensing machinery [270,271]. Rag GTPases directly bind to amino acids or their derivatives and relay that signal to mTORC1. Certain amino acids appear to be more essential for mTORC1 signaling than others; for example, leucine starvation inhibits mTORC1 and Sestrin2 [272,273]. Mechanistically, monomeric Sestrin2 binds to and antagonizes GATOR2 after leucine starvation, resulting in mTORC1 inhibition. Sestrin2 directly binds to leucine, leading to its dissociation from GATOR2 [274]. Similarly, the arginine sensor CASTOR1 directly binds to cytosolic arginine, inhibiting mTORC1 by its interaction with GATOR2 [275]. Interestingly, the sensor SLC38A9 allows arginine to interact with mTORC1 by transporting arginine-gated lysosomal amino acids, thereby enabling mTORC1 to sense both cytosolic and lysosomal arginine levels [276-278]. The amino acid transporters SLC1A5 and SLC7A5 in the plasma membrane supply cytosolic amino acids that activate mTORC1 signaling. In the absence of cytosolic amino acids, mTORC1 signaling is inhibited, leading to the activation of autophagy processes and protein degradation to replenish lysosomal amino acid pools [279,280]. Currently, it is unclear how mTORC1 activation is affected by other amino acids or the role of general amino acid sensors such as GCN2 and ATF4 in acute mTORC1 signaling cascades. While prolonged amino acid deprivation is thought to activate mTORC1 via GCN2 and ATF4, controlling the transcriptional upregulation by Sestrins, it is uncertain whether mTORC1 is regulated by GCN2 and ATF4 in transiently starved cells.
Conclusion And Future Prospective
Translational control is a key aspect of normal cell physiology, ensuring cellular survival under stress conditions. In this review, we have summarized the mechanisms of translation regulation at different stages during stress. Understanding stress-induced translational changes is vital for comprehending organismal homeostasis. This area of study, which encompasses mRNA translation efficiency, proteome adjustments, and molecular mechanisms, presents significant opportunities for discovery. Under stress conditions, non-canonical cap-dependent translation enables cells to adapt by translating specific stress response mRNAs, including those encoding survival proteins. Amid stress, modified translation fosters the synthesis of a distinct subset of proteins, potentially intersecting with longevity mechanisms and serving as targets for promoting healthy aging and combating diseases. Investigating these changes in the contexts of aging and disease holds clinical importance.
In the context of organismal aging, reducing translation errors is an effective strategy for increasing health span. Importantly, these discoveries contribute an additional layer to our existing comprehension of aging mechanisms. The modulation of protein synthesis, achieved through the reduction of initiation factors or ribosomal proteins, stands as a firmly established anti-aging intervention. The suggested underlying mechanisms for enhanced longevity encompass variations in translation processes. Pharmacological agents aimed at enhancing translation accuracy and potentially mitigating protein synthesis errors, particularly during the rate-limiting initiation phase, present promising avenues for addressing cancer, aging, and age-related ailments, notably neurodegenerative disorders primarily impacted by proteostasis decline. Currently, multiple drugs and antisense oligonucleotides are under evaluation against initiation factors to enhance cancer cell mortality. Given their ubiquity in translation processes, initiation factors hold considerable capacity for exploration in RNA-based therapeutics and chemical compound-based interventions for cancer treatment.
Several distinct translation-related regulatory stress responses are known, and the list is growing. Although recent years have seen substantial progress in characterizing new players in translational control, further examples remain to be identified. Research on protein translation deregulation relies on techniques like ribosome profiling, tRNA-sequencing to monitor and quantify different aspects of translation, mass spectrometry-based proteomics, and single-cell translation profiling and related processes have dramatically advanced this field. Each has its strengths and weaknesses, but combining them provides a fuller understanding. Advances in these methods, alongside other omics approaches, will deepen our knowledge. This integrated multidisciplinary approach will help develop better, personalized treatments, improving health outcomes, and combating diseases more effectively.
Acknowledgements
This work was supported by the grant from the NIH (R01AG062575-03S1). In addition, research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P20GM103423 and P20GM104318. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Conflict of Interest
None of the authors mentioned above have conflict of interest.
References
- Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34(3):146-153.
[Crossref] [Google Scholar] [PubMed]
- Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9(8):575-581.
[Crossref] [Google Scholar] [PubMed]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6(4):318-327.
[Crossref] [Google Scholar] [PubMed]
- Patel J, McLeod LE, Vries RG, Flynn A, Wang X, Proud CG. Cellular stresses profoundly inhibit protein synthesis and modulate the states of phosphorylation of multiple translation factors. Eur J Biochem. 2002;269(12):3076-3085.
[Crossref] [Google Scholar] [PubMed]
- Brito Querido J, Díaz-López I, Ramakrishnan V. The molecular basis of translation initiation and its regulation in eukaryotes. Nat Rev Mol Cell Biol. 2023:01-09.
[Crossref] [Google Scholar] [PubMed]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113-127.
[Crossref] [Google Scholar] [PubMed]
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19(6):568-576.
[Crossref] [Google Scholar] [PubMed]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40(2):228-237.
[Crossref] [Google Scholar] [PubMed]
- Merrick WC, Pavitt GD. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol. 2018;10(12):a033092.
[Crossref] [Google Scholar] [PubMed]
- Jennings MD, Kershaw CJ, Adomavicius T, Pavitt GD. Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes. Elife. 2017;6:e24542.
[Crossref] [Google Scholar] [PubMed]
- Sivan G, Elroy-Stein O. Regulation of mRNA Translation during cellular division. Cell cycle. 2008;7(6):741-744.
[Crossref] [Google Scholar] [PubMed]
- Leibovitch M, Topisirovic I. Dysregulation of mRNA translation and energy metabolism in cancer. Adv Biol Regul. 2018;67:30-39.
[Crossref] [Google Scholar] [PubMed]
- Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb Perspect Biol. 2012;4(10):a011544.
[Crossref] [Google Scholar] [PubMed]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, et al. mRNA helicases: The tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12(4):235-245.
[Crossref] [Google Scholar] [PubMed]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136(4):731-745.
[Crossref] [Google Scholar] [PubMed]
- Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: An overview. Cold Spring Harb Perspect Biol. 2012;4(12):a011528.
[Crossref] [Google Scholar] [PubMed]
- Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol. 2009;63:27-44.
[Crossref] [Google Scholar] [PubMed]
- Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283-331.
[Crossref] [Google Scholar] [PubMed]
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779-812.
[Crossref] [Google Scholar] [PubMed]
- Merrick WC. eIF4F: A retrospective. J Biol Chem. 2015;290(40):24091-24099.
[Crossref] [Google Scholar] [PubMed]
- Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5'-terminal cap in mRNA. Proc Natl Acad Sci. 1978;75(10):4843-4847.
[Crossref] [Google Scholar] [PubMed]
- Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, et al. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci. 2008;105(28):9564-9569.
[Crossref] [Google Scholar] [PubMed]
- Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39(6):2260-2270.
[Crossref] [Google Scholar] [PubMed]
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J Biol Chem. 2012;287(24):20301-20312.
[Crossref] [Google Scholar] [PubMed]
- Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467(7318):929-934.
[Crossref] [Google Scholar] [PubMed]
- Tavernarakis N. Protein synthesis and aging: eIF4E and the soma vs. germline distinction. Cell cycle. 2007;6(10):1168-1171.
[Crossref] [Google Scholar] [PubMed]
- Karamyshev AL, Patrick AE, Karamysheva ZN, Griesemer DS, Hudson H, Tjon-Kon-Sang S, et al. Inefficient SRP interaction with a nascent chain triggers an mRNA quality control pathway. Cell. 2014;156(1):146-157.
[Crossref] [Google Scholar] [PubMed]
- Pinarbasi ES, Karamyshev AL, Tikhonova EB, Wu IH, Hudson H, Thomas PJ. Pathogenic signal sequence mutations in progranulin disrupt SRP interactions required for mRNA stability. Cell reports. 2018;23(10):2844-2851.
[Crossref] [Google Scholar] [PubMed]
- Gregersen N, Bross P, Vang S, Christensen JH. Protein misfolding and human disease. Annu Rev Genomics Hum Genet.2006;7:103-124.
[Crossref] [Google Scholar] [PubMed]
- Hebert DN, Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87(4):1377-1408.
[Crossref] [Google Scholar] [PubMed]
- Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and-aggregation diseases. Trends Cell Biol. 2014;24(9):506-514.
[Crossref] [Google Scholar] [PubMed]
- Jarjanazi H, Savas S, Pabalan N, Dennis JW, Ozcelik H. Biological implications of SNPs in signal peptide domains of human proteins. Proteins. 2008;70(2):394-403.
[Crossref] [Google Scholar] [PubMed]
- Zimmermann R, Müller L, Wullich B. Protein transport into the endoplasmic reticulum: Mechanisms and pathologies. Trends Mol Med. 2006;12(12):567-573.
[Crossref] [Google Scholar] [PubMed]
- Hashimoto M, Rockenstein E, Crews L, Masliah E. Role of protein aggregation in mitochondrial dysfunction and neurodegeneration in Alzheimer’s and Parkinson’s diseases. Neuromolecular Med. 2003;4:21-35.
[Crossref] [Google Scholar] [PubMed]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(7):S10-S17.
[Crossref] [Google Scholar] [PubMed]
- Hipkiss AR. On why decreasing protein synthesis can increase lifespan. Mech Ageing Dev. 2007;128(5-6):412-414.
[Crossref] [Google Scholar] [PubMed]
- Mehta R, Chandler-Brown D, Ramos FJ, Shamieh LS, Kaeberlein M. Regulation of mRNA translation as a conserved mechanism of longevity control. Adv Exp Med Biol. 2010:14-29.
[Crossref] [Google Scholar] [PubMed]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18(4):564-570.
[Crossref] [Google Scholar] [PubMed]
- Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493(7432):338-345.
[Crossref] [Google Scholar] [PubMed]
- Beyer A, Hollunder J, Nasheuer HP, Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol Cell Proteomics. 2004;3(11):1083-1092.
[Crossref] [Google Scholar] [PubMed]
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:1-8.
[Crossref] [Google Scholar] [PubMed]
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966-3973.
[Crossref] [Google Scholar] [PubMed]
- Advani VM, Ivanov P. Translational control under stress: Reshaping the translatome. Bioessays. 2019;41(5):1900009.
[Crossref] [Google Scholar] [PubMed]
- Merrick WC, Pavitt GD. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol. 2018;10(12):a033092.
[Crossref] [Google Scholar] [PubMed]
- Wek RC. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol. 2018;10(7):a032870.
[Crossref] [Google Scholar] [PubMed]
- Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28(11):3729-3741.
[Crossref] [Google Scholar] [PubMed]
- Gatei M, Zhou BB, Hobson K, Scott S, Young D, Khanna KK. Ataxia Telangiectasia Mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites: In vivo assessment using phospho-specific antibodies. J Biol Chem. 2001;276(20):17276-17280.
[Crossref] [Google Scholar] [PubMed]
- Gehen SC, Staversky RJ, Bambara RA, Keng PC, O'Reilly MA. hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene. 2008;27(29):4065-74.
[Crossref] [Google Scholar] [PubMed]
- Gewandter JS, Bambara RA, o’Reilly MA. The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle. 2011;10(15):2561-2567.
[Crossref] [Google Scholar] [PubMed]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol. 1999;147(7):1431-1442.
[Crossref] [Google Scholar] [PubMed]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212(7).
[Crossref] [Google Scholar] [PubMed]
- Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849(7):861-870.
[Crossref] [Google Scholar] [PubMed]
- Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem Sci. 2013;38(10):494-506.
[Crossref] [Google Scholar] [PubMed]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871-884.
[Crossref] [Google Scholar] [PubMed]
- El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69-95.
[Crossref] [Google Scholar] [PubMed]
- Agris PF. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32(1):223-238.
[Crossref] [Google Scholar] [PubMed]
- Towns WL, Begley TJ. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: Activities, predications, and potential roles in human health. DNA Cell Biol. 2012;31(4):434-454.
[Crossref] [Google Scholar] [PubMed]
- Capra JD, Peterkofsky A. Effect of in vitro methylation on the chromatographic and coding properties of methyl-deficient leucine transfer RNA. J Mol Biol. 1968;33(3):591-607.
[Crossref] [Google Scholar] [PubMed]
- Karlsborn T, Tükenmez H, Mahmud AF, Xu F, Xu H, Byström AS. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol. 2014;11(12):1519-1528.
[Crossref] [Google Scholar] [PubMed]
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nature communications. 2012 Jan;3(1):937.
[Crossref] [Google Scholar] [PubMed]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639-2649.
- Sun C, Fu Z, Wang S, Li J, Li Y, Zhang Y, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16-25.
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407-450.
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6(2):269-279.
- Rousakis A, Vlassis A, Vlanti A, Patera S, Thireos G, Syntichaki P. The general control nonderepressible‐2 kinase mediates stress response and longevity induced by target of rapamycin inactivation in Caenorhabditis elegans. Aging Cell. 2013;12(5):742-751.
- Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 2003;17(7):859-872.
- Cuesta R, Laroia G, Schneider RJ. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14(12):1460-1470.
[Google Scholar] [PubMed]
- Ma S, Bhattacharjee RB, Bag J. Expression of poly (A)‐binding protein is upregulated during recovery from heat shock in HeLa cells. The FEBS journal. 2009;276(2):552-570.
- Datu AK, Bag J. Enhanced translation of mRNAs encoding proteins involved in mRNA translation during recovery from heat shock. PLoS One. 2013;8(5):e64171.
- Burgess HM, Gray NK. An integrated model for the nucleo-cytoplasmic transport of cytoplasmic poly (A)-binding proteins. Commun Integr Biol. 2012;5(3):243-247.
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5(10):827-835.
- Trinh MA, Klann E. Translational control by eIF2α kinases in long-lasting synaptic plasticity and long-term memory. Neurobiol Learn Mem. 2013;105:93-99.
- Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly (A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol.2006;71:537-543.
- Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly (A)‐binding protein stimulate translation in vivo. EMBO J. 2000.
- Kahvejian A, Roy G, Sonenberg N. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol. 2001;66:293-300.
- Leppek K, Das R, Barna M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19(3):158-174.
- Pelletier JE, Sonenberg NA. Internal binding of eukaryotic ribosomes on poliovirus RNA: Translation in HeLa cell extracts. J Virol. 1989;63(1):441-444.
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113-127.
- Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1(2):253-265.
- Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1-1.
- Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39(4):197-216.
- Fischer PM. Cap in hand: Targeting eIF4E. Cell cycle. 2009;8(16):2535-2541.
- Goodfellow IG, Roberts LO. Eukaryotic initiation factor 4E. ThInt J Biochem Cell Biol. 2008;40(12):2675-2680.
- Arribas-Layton M, Wu D, Lykke-Andersen J, Song H. Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim Biophys Acta. 2013;1829(6-7):580-589.
- Kneller EL, Rakotondrafara AM, Miller WA. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119(1):63-75.
- Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol. 2019;11(3):a033001.
- Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, et al. Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol Cell Biol. 2012;32(19):3978-3989.
- Villa N, Do A, Hershey JW, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c,-d, and-e to promote mRNA recruitment to the ribosome. J Biol Chem. 2013;288(46):32932-32940.
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, et al. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016;351(6270):aad4939.
[Crossref] [Google Scholar] [PubMed]
- Sokabe M, Fraser CS. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc Natl Acad Sci. 2017;114(24):6304-6309.
[Crossref] [Google Scholar] [PubMed]
- Andreev DE, O’Connor PB, Zhdanov AV, Dmitriev RI, Shatsky IN, Papkovsky DB, et al. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 2015;16:1-4.
[Crossref] [Google Scholar] [PubMed]
- Ho JD, Lee S. A cap for every occasion: Alternative eIF4F complexes. Trends Biochem Sci. 2016;41(10):821-823.
[Crossref] [Google Scholar] [PubMed]
- Shatsky IN, Dmitriev SE, Andreev DE, Terenin IM. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit Rev Biochem Mol Biol. 2014;49(2):164-177.
[Crossref] [Google Scholar] [PubMed]
- Ali MU, Ur Rahman MS, Jia Z, Jiang C. Eukaryotic translation initiation factors and cancer. Tumour Biol. 2017;39(6):1010428317709805.
[Crossref] [Google Scholar] [PubMed]
- Yang X, Zhong W, Cao R. Phosphorylation of the mRNA cap-binding protein eIF4E and cancer. Cell Signal. 2020;73:109689.
[Crossref] [Google Scholar] [PubMed]
- Karaki S, Andrieu C, Ziouziou H, Rocchi P. The eukaryotic translation initiation factor 4E (eIF4E) as a therapeutic target for cancer. Adv Protein Chem Struct Biol. 2015;101:01-26.
[Crossref] [Google Scholar] [PubMed]
- Lukhele S, Bah A, Lin H, Sonenberg N, Forman-Kay JD. Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure. 2013;21(12):2186-2196.
[Crossref] [Google Scholar] [PubMed]
- Paku KS, Umenaga Y, Usui T, Fukuyo A, Mizuno A, In Y, et al. A conserved motif within the flexible C-terminus of the translational regulator 4E-BP is required for tight binding to the mRNA cap-binding protein eIF4E. Biochem J. 2012;441(1):237-245.
[Crossref] [Google Scholar] [PubMed]
- Peter D, Igreja C, Weber R, Wohlbold L, Weiler C, Ebertsch L, et al. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol Cell. 2015;57(6):1074-1087.
[Crossref] [Google Scholar] [PubMed]
- Nandagopal N, Roux PP. Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation. 2015;3(1):e983402.
[Crossref] [Google Scholar] [PubMed]
- Chen X, An Y, Tan M, Xie D, Liu L, Xu B. Biological functions and research progress of eIF4E. Front Oncol. 2023;13.
[Crossref] [Google Scholar] [PubMed]
- Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21(24):3232-3237.
[Crossref] [Google Scholar] [PubMed]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci. 2010;107(32):14134-14139.
[Crossref] [Google Scholar] [PubMed]
- Martinez A, Sese M, Losa JH, Robichaud N, Sonenberg N, Aasen T, et al. Phosphorylation of eIF4E confers resistance to cellular stress and DNA-damaging agents through an interaction with 4E-T: A rationale for novel therapeutic approaches. PLoS One. 2015;10(4):e0123352.
[Crossref] [Google Scholar] [PubMed]
- Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175(3):415-426.
[Crossref] [Google Scholar] [PubMed]
- Nishimura T, Padamsi Z, Fakim H, Milette S, Dunham WH, Gingras AC, et al. The eIF4E-binding protein 4E-T is a component of the mRNA decay machinery that bridges the 5′ and 3′ termini of target mRNAs. Cell reports. 2015;11(9):1425-1436.
[Crossref] [Google Scholar] [PubMed]
- Osborne MJ, Borden KL. The eukaryotic translation initiation factor eIF 4E in the nucleus: Taking the road less traveled. Immunol Rev. 2015;263(1):210-223.
[Crossref] [Google Scholar] [PubMed]
- Sesma A, Castresana C, Castellano MM. Regulation of translation by TOR, eIF4E and eIF2α in plants: Current knowledge, challenges and future perspectives. Front Plant Sci. 2017;8:644.
[Crossref] [Google Scholar] [PubMed]
- Topisirovic I, Siddiqui N, Lapointe VL, Trost M, Thibault P, Bangeranye C, et al. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export‐competent RNP. EMBO J. 2009;28(8):1087-1098.
[Crossref] [Google Scholar] [PubMed]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134(6):1042-1054.
[Crossref] [Google Scholar] [PubMed]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477-480.
[Crossref] [Google Scholar] [PubMed]
- Jung MY, Lorenz L, Richter JD. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol Cell Biol. 2006;26(11):4277-4287.
[Crossref] [Google Scholar] [PubMed]
- Rhoads RE. eIF4E: New family members, new binding partners, new roles. J Biol Chem. 2009;284(25):16711-16715.
[Crossref] [Google Scholar] [PubMed]
- Wells DG. RNA-binding proteins: A lesson in repression. J Neurosci. 2006;26(27):7135-7138.
[Crossref] [Google Scholar] [PubMed]
- Dever TE, Kinzy TG, Pavitt GD. Mechanism and regulation of protein synthesis in Saccharomyces cerevisiae. Genetics. 2016;203(1):65-107.
[Crossref] [Google Scholar] [PubMed]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva CG, et al. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159(3):597-607.
[Crossref] [Google Scholar] [PubMed]
- Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell. 2015;59(3):399-412.
[Crossref] [Google Scholar] [PubMed]
- Hinnebusch AG. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci. 2017;42(8):589-611.
[Crossref] [Google Scholar] [PubMed]
- Bogorad AM, Lin KY, Marintchev A. Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res. 2017;45(20):11962-11979.
[Crossref] [Google Scholar] [PubMed]
- Majumder M, Mitchell D, Merkulov S, Wu J, Guan BJ, Snider MD, et al. Residues required for phosphorylation of translation initiation factor eIF2α under diverse stress conditions are divergent between yeast and human. Int J Biochem Cell Biol. 2015;59:135-141.
[Crossref] [Google Scholar] [PubMed]
- Castilho BA, Shanmugam R, Silva RC, Ramesh R, Himme BM, Sattlegger E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim Biophys Acta. 2014;1843(9):1948-1968.
[Crossref] [Google Scholar] [PubMed]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, et al. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999;13(11):1422-1437.
[Crossref] [Google Scholar] [PubMed]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 2012;8(6):e1002760.
[Crossref] [Google Scholar] [PubMed]
- Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci. 2013;70:3493-3511.
[Crossref] [Google Scholar] [PubMed]
- Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: A review. Diabetes. 2002;51(suppl_3):S455-S461.
[Crossref] [Google Scholar] [PubMed]
- Simpson CE, Ashe MP. Adaptation to stress in yeast: To translate or not? Biochem Soc Trans. 2012;40(4):794-799.
[Crossref] [Google Scholar] [PubMed]
- Kong J, Lasko P. Translational control in cellular and developmental processes. Nat Rev Genet. 2012;13(6):383-394.
[Crossref] [Google Scholar] [PubMed]
- Ayala A, Parrado J, Bougria M, Machado A. Effect of oxidative stress, produced by cumene hydroperoxide, on the various steps of protein synthesis: Modifications of elongation factor-2. J Biol Chem. 1996;271(38):23105-23110.
[Crossref] [Google Scholar] [PubMed]
- Kang KR, Lee SY. Effect of serum and hydrogen peroxide on the Ca2+/calmodulin-dependent phosphorylation of eukaryotic elongation factor 2 (eEF-2) in Chinese hamster ovary cells. Exp Mol Med. 2001;33(4):198-204.
[Crossref] [Google Scholar] [PubMed]
- Piwien-Pilipuk G, Ayala A, Machado A, Galigniana MD. Impairment of Mineralocorticoid Receptor (MR)-dependent biological response by oxidative stress and aging: Correlation with post-translational modification of MR and decreased ADP-ribosylatable level of elongation factor 2 in kidney cells. J Biol Chem. 2002;277(14):11896-11903.
[Crossref] [Google Scholar] [PubMed]
- Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, et al. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281(39):29011-29021.
[Crossref] [Google Scholar] [PubMed]
- Ryazanov AG, Pavur KS, Dorovkov MV. Alpha-kinases: A new class of protein kinases with a novel catalytic domain. Curr Biol. 1999;9(2):R43-R45.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Regufe da Mota S, Liu R, Moore CE, Xie J, Lanucara F, et al. Eukaryotic elongation factor 2 kinase activity is controlled by multiple inputs from oncogenic signaling. Mol Cell Biol. 2014;34(22):4088-4103.
[Crossref] [Google Scholar] [PubMed]
- Kenney JW, Moore CE, Wang X, Proud CG. Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul. 2014;55:15-27.
[Crossref] [Google Scholar] [PubMed]
- Shetty S, Hofstetter J, Battaglioni S, Ritz D, Hall MN. TORC1 phosphorylates and inhibits the ribosome preservation factor Stm1 to activate dormant ribosomes. EMBO J. 2023;42(5):e112344.
[Crossref] [Google Scholar] [PubMed]
- Chen PJ, Huang YS. CPEB2-eEF2 interaction impedes HIF‐1α RNA translation. EMBO J. 2012;31(4):959-971.
[Crossref] [Google Scholar] [PubMed]
- Kruiswijk F, Yuniati L, Magliozzi R, Low TY, Lim R, Bolder R, et al. Coupled activation and degradation of eEF2K regulates protein synthesis in response to genotoxic stress. Sci Signal. 2012;5(227):40.
[Crossref] [Google Scholar] [PubMed]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21(4):521-531.
[Crossref] [Google Scholar] [PubMed]
- Pelechano V, Wei W, Steinmetz LM. Widespread co-translational RNA decay reveals ribosome dynamics. Cell. 2015;161(6):1400-1412.
[Crossref] [Google Scholar] [PubMed]
- Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, et al. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem. 2009;284(37):25254-25267.
[Crossref] [Google Scholar] [PubMed]
- Chen CW, Tanaka M. Genome-wide translation profiling by ribosome-bound tRNA capture. Cell Rep. 2018;23(2):608-621.
[Crossref] [Google Scholar] [PubMed]
- Roy R, Rajyaguru PI. Stress granules and p-bodies: An insight into mRNA translational control and decay. 2018.
- Decker CJ, Parker R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4(9):a012286.
[Crossref] [Google Scholar] [PubMed]
- Luo Y, Na Z, Slavoff SA. P-bodies: Composition, properties, and functions. Biochemistry. 2018;57(17):2424-2431.
[Crossref] [Google Scholar] [PubMed]
- Mahboubi H, Stochaj U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim Biophys Acta Mol Basis Dis. 2017;1863(4):884-895.
[Crossref] [Google Scholar] [PubMed]
- Grousl T, Ivanov P, Frydlová I, Vasicová P, Janda F, Vojtová J, et al. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122(12):2078-2088.
[Crossref] [Google Scholar] [PubMed]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729-1732.
[Crossref] [Google Scholar] [PubMed]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753-767.
[Crossref] [Google Scholar] [PubMed]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336-340.
[Crossref] [Google Scholar] [PubMed]
- Hubstenberger A, Noble SL, Cameron C, Evans TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell. 2013;27(2):161-173.
[Crossref] [Google Scholar] [PubMed]
- Hubstenberger A, Cameron C, Noble SL, Keenan S, Evans TC. Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J Cell Biol. 2015;211(3):703-716.
[Crossref] [Google Scholar] [PubMed]
- Iwaki A, Izawa S. Acidic stress induces the formation of P-bodies, but not stress granules, with mild attenuation of bulk translation in Saccharomyces cerevisiae. Biochem J. 2012;446(2):225-233.
[Crossref] [Google Scholar] [PubMed]
- Takahashi S, Sakurai K, Ebihara A, Kajiho H, Saito K, Kontani K, et al. RhoA activation participates in rearrangement of processing bodies and release of nucleated AU-rich mRNAs. Nucleic Acids Res. 2011;39(8):3446-3457.
[Crossref] [Google Scholar] [PubMed]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300(5620):805-808.
[Crossref] [Google Scholar] [PubMed]
- Aizer A, Kalo A, Kafri P, Shraga A, Ben-Yishay R, Jacob A, et al. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J Cell Sciscience. 2014;127(20):4443-4456.
[Crossref] [Google Scholar] [PubMed]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310(5747):486-489.
[Crossref] [Google Scholar] [PubMed]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell. 2017;68(1):144-157.
[Crossref] [Google Scholar] [PubMed]
- Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014;11(8):1019-1030.
[Crossref] [Google Scholar] [PubMed]
- Ayache J, Bénard M, Ernoult-Lange M, Minshall N, Standart N, Kress M, et al. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol Biol Cell. 2015;26(14):2579-2595.
[Crossref] [Google Scholar] [PubMed]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007.
[Crossref] [Google Scholar] [PubMed]
- Stoecklin G, Mayo T, Anderson P. ARE‐mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7(1):72-77.
[Crossref] [Google Scholar] [PubMed]
- Blake LA, Watkins L, Liu Y, Inoue T, Wu B. A rapid inducible RNA decay system reveals fast mRNA decay in P-bodies. Nat Commun. 2024;15(1):2720.
[Crossref] [Google Scholar] [PubMed]
- Adjibade P, Mazroui R. Stress granules: Stress-induced cytoplasmic mRNPs compartments linked to mRNA translational regulatory pathways. Front RNA Res. 2023;1:1226610.
- Riggs CL, Kedersha N, Ivanov P, Anderson P. Mammalian stress granules and P bodies at a glance. J Cell Sci. 2020;133(16):242487.
[Crossref] [Google Scholar] [PubMed]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284(2):C273-C284.
[Crossref] [Google Scholar] [PubMed]
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285-298.
[Crossref] [Google Scholar] [PubMed]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164(3):487-498.
[Crossref] [Google Scholar] [PubMed]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040-1044.
[Crossref] [Google Scholar] [PubMed]
- Kedersha NA, Anderson P. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30(6):963-969.
[Crossref] [Google Scholar] [PubMed]
- Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell. 2018;174(4):791-802.
[Crossref] [Google Scholar] [PubMed]
- Protter DS, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668-679.
[Crossref] [Google Scholar] [PubMed]
- Van Treeck B, Parker R. Principles of stress granules revealed by imaging approaches. Cold Spring Harb Perspect Biol. 2019;11(2):a033068.
[Crossref] [Google Scholar] [PubMed]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118(5):981-992.
[Crossref] [Google Scholar] [PubMed]
- Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63(5):796-810.
[Crossref] [Google Scholar] [PubMed]
- Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995.
[Crossref] [Google Scholar] [PubMed]
- Srivastava SP, Kumar KU, Kaufman RJ. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J Biol Chem. 1998;273(4):2416-2423.
[Crossref] [Google Scholar] [PubMed]
- McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, et al. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280(17):16925-16933.
[Crossref] [Google Scholar] [PubMed]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife. 2013;2:e00498.
[Crossref] [Google Scholar] [PubMed]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife. 2015;4:e05033.
[Crossref] [Google Scholar] [PubMed]
- Nabiałek E, Wańha W, Kula D, Jadczyk T, Krajewska M, Kowalówka A, et al. In acute myocardial infarction and stable coronary heart disease. Minerva Cardioangiol. 2013;61:627-637.
[Google Scholar] [PubMed]
- Fukao A, Aoyama T, Fujiwara T. The molecular mechanism of translational control via the communication between the microRNA pathway and RNA-binding proteins. RNA Biol. 2015;12(9):922-926.
[Crossref] [Google Scholar] [PubMed]
- Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007(367):1.
[Crossref] [Google Scholar] [PubMed]
- Kim VN. MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376-385.
[Crossref] [Google Scholar] [PubMed]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509-524.
[Crossref] [Google Scholar] [PubMed]
- O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:388354.
[Crossref] [Google Scholar] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell. 2005;120(1):15-20.
[Crossref] [Google Scholar] [PubMed]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642-655.
[Crossref] [Google Scholar] [PubMed]
- Akgül B, Erdoğan İ. Intracytoplasmic re-localization of miRISC complexes. Front Genet. 2018;9:417037.
[Crossref] [Google Scholar] [PubMed]
- Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE. Pairing beyond the seed supports microRNA targeting specificity. Mol Cell. 2016;64(2):320-333.
[Crossref] [Google Scholar] [PubMed]
- Makarova JA, Shkurnikov MU, Wicklein D, Lange T, Samatov TR, Turchinovich AA, et al. Intracellular and extracellular microRNA: An update on localization and biological role. Prog Histochem Cytochem. 2016;51(3-4):33-49.
[Crossref] [Google Scholar] [PubMed]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25(5):635-646.
[Crossref] [Google Scholar] [PubMed]
- Anderson P, Kedersha N. Stress granules: The Tao of RNA triage. Trends Biochem Sci. 2008;33(3):141-150.
[Crossref] [Google Scholar] [PubMed]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci. 2006;103(48):18125-18130.
[Crossref] [Google Scholar] [PubMed]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99-110.
[Crossref] [Google Scholar] [PubMed]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 2006;20(14):1885-1898.
[Crossref] [Google Scholar] [PubMed]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277(5322):99-101.
[Crossref] [Google Scholar] [PubMed]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, et al. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18(11):1218-1226.
[Crossref] [Google Scholar] [PubMed]
- Ding J, Zhou S, Guan J. Finding microRNA targets in plants: Current status and perspectives. Genomics Proteomics Bioinformatics. 2012;10(5):264-275.
[Crossref] [Google Scholar] [PubMed]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594-596.
[Crossref] [Google Scholar] [PubMed]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54(5):751-765.
[Crossref] [Google Scholar] [PubMed]
- Vaidya AT, Rente SR, Pratt GA, Prasad A, Schoenberg DR, Parker R. Regulation of translation initiation by the CCR4-NOT deadenylase complex. Biochem. 2021;60(34):2619-2629
- Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, et al. mRNA deadenylation is coupled to translation rates by the CCR4-NOT complex. Nature. 2022;592(7852):624-629.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight?. Nat Rev Genet. 2008;9(2):102-114.
[Crossref] [Google Scholar] [PubMed]
- Wu L, Belasco JG. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Fabbri M. MicroRNAs and miRceptors: A new mechanism of action for intercellular communication. Philos Trans R Soc Lond B Biol Sci. 2018;373(1737):20160486.
[Crossref] [Google Scholar] [PubMed]
- Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 2019;30(4):656-673.
[Crossref] [Google Scholar] [PubMed]
- Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9(2):276.
[Crossref] [Google Scholar] [PubMed]
- Mansoori B, Baradaran B, Nazari A, Gaballu FA, Cho WC, Mansoori B. MicroRNAs in the cancer cell-to-cell communication: An insight into biological vehicles. Biomed Pharmacother. 2022;153:113449.
[Crossref] [Google Scholar] [PubMed]
- Levantini E, Rizzo M. miRNAs: From master regulators of gene expression to biomarkers involved in intercellular communication. Biomedicines. 2024;12(4):721.
[Crossref] [Google Scholar] [PubMed]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485(7396):109-113.
[Crossref] [Google Scholar] [PubMed]
- Khoutorsky A, Bonin RP, Sorge RE, Gkogkas CG, Pawlowski SA, Jafarnejad SM, et al. Translational control of nociception via 4E-binding protein 1. Elife. 2015;4:e12002.
[Crossref] [Google Scholar] [PubMed]
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960-976.
[Crossref] [Google Scholar] [PubMed]
- DuRose JB, Scheuner D, Kaufman RJ, Rothblum LI, Niwa M. Phosphorylation of eukaryotic translation initiation factor 2α coordinates rRNA transcription and translation inhibition during endoplasmic reticulum stress. Mol Cell Biol. 2009;29(15):4295-4307.
[Crossref] [Google Scholar] [PubMed]
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23(23):8862-8877.
[Crossref] [Google Scholar] [PubMed]
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15(9):4990-4997.
[Crossref] [Google Scholar] [PubMed]
- Philippe L, van den Elzen AM, Watson MJ, Thoreen CC. Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5′ TOP motifs. Proc Natl Acad Sci. 2020;117(10):5319-5328.
[Crossref] [Google Scholar] [PubMed]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, et al. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24(8):3112-3124.
[Crossref] [Google Scholar] [PubMed]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123(4):569-580.
[Crossref] [Google Scholar] [PubMed]
- Yi YW, You KS, Park JS, Lee SG, Seong YS. Ribosomal protein S6: A potential therapeutic target against cancer?. Int J Mol Sci. 2021;23(1):48.
[Crossref] [Google Scholar] [PubMed]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO. 2006;25(12):2781-2791.
[Crossref] [Google Scholar] [PubMed]
- Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta. 2015;1849(7):801-811.
[Crossref] [Google Scholar] [PubMed]
- Nakagawa T, Araki T, Nakagawa M, Hirao A, Unno M, Nakayama K. S6 kinase-and β-TrCP2-dependent degradation of p19Arf is required for cell proliferation. Mol Cell Biol. 2015;35(20):3517-27.
[Crossref] [Google Scholar] [PubMed]
- Wang Q, Yang HS. The role of Pdcd4 in tumour suppression and protein translation. Biol Cell. 2018;110(8):169-177.
[Crossref] [Google Scholar] [PubMed]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1-and ßTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314(5798):467-471.
[Crossref] [Google Scholar] [PubMed]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence Jr JC. mTOR‐dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO. 2006;25(8):1659-1668.
[Crossref] [Google Scholar] [PubMed]
- Chaillou T, Kirby TJ, McCarthy JJ. Ribosome biogenesis: Emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol. 2014;229(11):1584-1594.
[Crossref] [Google Scholar] [PubMed]
- Tsang CK, Liu H, Zheng XS. mTOR binds to the promoters of RNA polymerase I-and III-transcribed genes. Cell Cycle. 2010;9(5):953-957.
[Crossref] [Google Scholar] [PubMed]
- Von Walden F, Liu C, Aurigemma N, Nader GA. mTOR signaling regulates myotube hypertrophy by modulating protein synthesis, rDNA transcription, and chromatin remodeling. Am J Physiol Cell Physiol. 2016;311(4):C663-C672.
[Crossref] [Google Scholar] [PubMed]
- Schnapp A, Pfleiderer C, Rosenbauer H, Grummt I. A growth‐dependent transcription initiation factor (TIF‐IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO. 1990;9(9):2857-2863.
[Crossref] [Google Scholar] [PubMed]
- Damgaard CK, Lykke-Andersen J. Translational coregulation of 5′ TOP mRNAs by TIA-1 and TIAR. Genes Dev. 2011;25(19):2057-2068.
[Crossref] [Google Scholar] [PubMed]
- Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18(4):423-434.
[Crossref] [Google Scholar] [PubMed]
- Yang K, Yang J, Yi J. Nucleolar stress: Hallmarks, sensing mechanism and diseases. Cell stress. 2018;2(6):125.
[Crossref] [Google Scholar] [PubMed]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437-440.
[Crossref] [Google Scholar] [PubMed]
- McIntosh KB, Bonham-Smith PC. Ribosomal protein gene regulation: What about plants? Botany. 2006;84(3):342-362.
- Moss T, Stefanovsky VY. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase. Prog Nucleic Acid Res Mol Biol. 1995;50:25-66.
[Crossref] [Google Scholar] [PubMed]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7(3):303-310.
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7(3):311-318.
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471-484.
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139(1):149-160.
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, et al. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14(1):55-66.
- Takauji Y, Wada T, Takeda A, Kudo I, Miki K, Fujii M, et al. Restriction of protein synthesis abolishes senescence features at cellular and organismal levels. Sci Rep. 2016;6(1):18722.
- Ren X, Hu B, Song M, Ding Z, Dang Y, Liu Z, et al. Maintenance of nucleolar homeostasis by CBX4 alleviates senescence and osteoarthritis. Cell Rep. 2019;26(13):3643-3656.
- Kim HS, Pickering AM. Protein translation paradox: Implications in translational regulation of aging. Front cell dev biol. 2023;11:1129281.
- Howard AC, Mir D, Snow S, Horrocks J, Sayed H, Ma Z, et al. Anabolic function downstream of TOR controls trade-offs between longevity and reproduction at the level of specific tissues in C. elegans. Front aging. 2021;2:725068.
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging cell. 2007;6(1):95-110.
- Kaeberlein M, Powers III RW, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310(5751):1193-1196.
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140-144.
- Shore DE, Ruvkun G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 2013;23(9):409-420.
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133(2):292-302.
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42(4):275-286.
- Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes and development. 2003;17(16):2006-2020.
- Hsieh CC, Papaconstantinou J. Akt/PKB and p38 MAPK signaling, translational initiation and longevity in Snell dwarf mouse livers. Mech Ageing Dev. 2004;125(10-11):785-798.
- Sharp ZD, Bartke A. Evidence for down-regulation of phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in Ames dwarf mice. J Gerontol A Biol Sci Med Sci. 2005;60(3):293-300.
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging cell. 2007;6(1):111-119.
- Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445(7130):922-926.
- Johnson JE, Johnson FB. Methionine restriction activates the retrograde response and confers both stress tolerance and lifespan extension to yeast, mouse and human cells. PloS one. 2014;9(5):e97729.
- Shu XE, Swanda RV, Qian SB. Nutrient control of mRNA translation. Annu Rev Nutr. 2020;40(1):51-75.
- Gameiro PA, Struhl K. Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep. 2018;24(6):1415-1424.
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273(23):14484-14494.
- Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, et al. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2004;18(11):1303.
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci. 2007;104(49):19345-19350.
- Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer cell. 2011;19(5):613-628.
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, et al. Serine starvation induces stress and p53-dependent metabolic remodeling in cancer cells. Nature. 2013;493(7433):542-546.
- Edwards C, Canfield J, Copes N, Brito A, Rehan M, Lipps D, et al. Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans. BMC Genet. 2015; 16(1):1-24.
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009; 462(7276):1061-1064.
- Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993; 123(2):269-274.
- Richie Jr JP, Leutzinger Y, Parthasarathy S, Maixoy V, Orentreich N, Zimmerman JA. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8(15):1302-1307.
- Kim J, Guan KL. mTOR as a central hub of nutrient signaling and cell growth. Nat Cell Biol. 2019; 21(1):63-71.
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274-293.
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008; 10(8):935-945.
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008; 320(5882):1496-1501.
- Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin-sensitive pathway. J Nutr. 2000;130(10):2413-2419.
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016; 351(6268):43-48.
- Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536(7615):229-233.
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165(1):153-164.
- Jung J, Genau HM, Behrends C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol Cell Biol. 2015;35(14):2479-2494.
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347(6218):188-194.
- Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171(3):642-654.
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981-1991.
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132-141.
Citation: Mir DA, Ma Z, Horrocks J, Rogers A (2024) Stress-Induced Eukaryotic Translational Regulatory Mechanisms. J Clin Med Sci. 8:277.
Copyright: © 2024 Mir DA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
