Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2015) Volume 6, Issue 5
Specific Detection of Klebsiella variicola and K. oxytoca by Loop-Mediated Isothermal Amplification
Abstract
Klebsiella spp. are opportunistic pathogens with clinical, veterinary and plant-associated isolates. A previous study showed that bacterial ooze from wetwood of severely declined ironwood trees in Guam contained Ralstonia solanacearum, Klebsiella variicola and K. oxytoca. In this study, Loop-Mediated Isothermal Amplification (LAMP) detected K. variicola and K. oxytoca specifically, using unique primer sets designed individually for each organism. Each LAMP detected its target specifically, while showing negative results for non-target bacteria and negative controls. LAMP detected Klebsiella in inoculated-ironwood stem tissues and bacterial ooze. Due to the presence of plant inhibitors, different sampling protocols were tested. Soaking plant tissue samples to allow diffusion of bacteria into solution, followed by boiling, provided optimum detection of Klebsiella directly from plant samples. False negatives obtained when using crushed plant samples were eliminated by including an enrichment step, involving plating and 12-h incubation. DGGE (denaturing gradient gel electrophoresis) and a colony-blot immunoassay using a Klebsiella-specific antibody also detected Klebsiella in inoculated ironwood. DGGE bands and antibody crossreactions from closely related enterobacters showed the potential for false positive results. The nature of LAMP makes it ideal for point-of-care testing, and when combined with the specificity of the LAMP primers developed in this study, demonstrates its potential as a routine field test for Klebsiella in ironwood in Guam, as well as clinical and veterinary diagnosis of Klebsiella infection. Additionally, the regions targeted for detection in this study have application across all forms of molecular-based diagnostics.
Keywords: Klebsiella; Detection; Loop-mediated amplification; Casuarina equisetifolia; ironwood; Decline
Introduction
Ironwood (Casuarina equisetifolia) is a common forest tree in the Pacific islands and is widely propagated in Australia, India and China for firewood, wood for construction, land reclamation and windbreaks. Ironwood is salt tolerant and grows on nutrient-poor sandy soils common on coastlines [1]. For the past twelve years, ironwood trees on Guam have been undergoing a slow decline and thousands have died throughout the island [2-4]. Ironwood decline is associated with thinning foliage, dieback of branches, and wetwood formation. Various causal agents, including the root and butt rot fungus Ganoderma spp., termites, tree-care practices, and most recently bacteria, have been implicated [5]. However, the disease etiology has not been clearly established [2,4,6,7]. A disease showing similar symptoms was first reported in Mauritius [8], and later in India [9]. The disease symptoms include defoliation, yellowing, wilt, dieback and finally death of the trees.
In a 2011 survey of ironwood decline in Guam, involvement of Ralstonia solanacearum (Rs), a bacterial pathogen that causes wilt in a large number of plant hosts including ironwood, was suspected when ooze from diseased trees produced positive reactions with Rsspecific immunostrips (Agdia, Inc.). Following the initial observation by Putnam in 2011 [10], numerous samples from symptomatic plants were sent to Hawai‘i for isolation and identification. However, Rs could not be cultured, resulting in an additional survey in Guam in 2012. Three complimentary approaches were used to isolate and identify the pathogen: growth on modified Rs-semiselective medium (mSMSA), Rs-specific immunostrip assays, and confirmation with a Rs-specific loop-mediated isothermal amplification (LAMP) reaction [11,12]. The identity of isolates was determined in Hawai‘i and the two predominant types of bacteria associated with ironwood decline were characterized [13,14]. Bacteriological tests, along with multiplex PCR, BOX-PCR, and phylogenetic analysis of the dnaA RIF marker [15], were performed to further characterize and evaluate the genetic diversity among the Rs isolates [13,14]. Non-Rs isolates were presumptively identified as Klebsiella spp. using bacteriological tests [13,14,16,17]. Most strains showed closest similarity to K. variicola following sequence analysis of the 16S rRNA gene, while one showed closest match to K. oxytoca. Strains identified as Rs were capable of wilting tomato seedlings within 7-10 days and ironwood seedlings within 10-30 days. Klebsiella strains did not produce symptoms in inoculated plants, but were re-isolated from stem tissues as far as 10 cm above the inoculation site, indicating possible colonization of the plant. Results indicated, for the first time, that Rs is involved in the decline of ironwood in Guam and Klebsiella spp. are associated with wetwood, a phenomenon associated with various bacteria and reported in other forest ecosystems [18,19].
Ironwood decline has not yet been observed in Hawai‘i, or even Saipan, which is much closer to Guam, even though Rs is present throughout the Pacific. Molecular and immunodiagnostic tools for rapid pathogen identification are needed to obtain relevant data in future field studies to better understand ironwood decline and its potential to spread to other islands of the Pacific. Immunostrips (Agdia, Inc.) and LAMP technology using a hand-held SMART-DART device [11,12] are available for Rs detection, but similar tests are needed to determine the possible role of Klebsiella spp. in ironwood decline. Once developed and validated, fieldready diagnostic tools can be used to determine the association of these two bacteria within naturally infected symptomatic plants, and lack of association with healthy ones. These assays could also be used to screen nursery trees for latent and active infections, thereby providing tools to produce healthy nursery stocks for the replenishment of dead ironwood trees and reforestation in Guam. This study compares several diagnostic approaches, including a LAMP assay, for detecting Klebsiella spp. in ironwood, as well as mixed cultures with Ralstonia solanacearum and other bacterial genera
Materials and Methods
Bacterial strains and culture conditions
The strains used in this study (Table 1) were from the Pacific Bacterial Collection at the University of Hawai’i at Mānoa and included three Klebsiella variicola strains (A6126, A6127, and A6128), one K. oxytoca strain (A6125) and twoRalstonia solanacearum strains (A6123 and A6124), which were all isolated from ironwood in Guam, one K. pneumoniae strain (A6133, aka ATCC 13883), as well as Agrobacterium tumefaciens (A2961, aka C58), Enterobacter aerogenes (A3131, aka ATCC 13048), E. cloacae (A5149), Escherichia coli (A6091, aka LJH 1947), Pseudomonas fluorescens (A3275, aka A811-1), and Ralstonia solanacearum (GMI 1000). Bacteria were removed from storage, plated onto TZC medium (17 g/L agar, 10 g/L peptone, 5 g/L glucose and 0.001% 2,3,5-triphenyl-tetrazolium chloride (TZC)) and then incubated at 26°C (±2°C).
| Organism | Strain | Other ID | Hcp 1 LAMP | UGH LAMP |
|---|---|---|---|---|
| Agrobacterium tumefaciens | A2961 | C58 | - | - |
| Enterobacteraerogenes | A3133 | ATCC 13048 | - | - |
| Enterobacter cloacae | A5149 | B193 | - | - |
| Escherichia coli | A6091 | LJH 1947 | - | - |
| Klebsiella oxytoca | A6125 | S-3 | - | + |
| Klebsiella pneumoniae | A6133 | ATCC 13883 | - | - |
| Klebsiella variicola | A6126 | S-4 | + | - |
| Klebsiella variicola | A6127 | S-8 | + | - |
| Klebsiella variicola | A6128 | S-19 | + | - |
| Pseudomonas fluorescens | A3275 | A811-1 | - | - |
| Ralstoniasolanacearum | A6123 | S-25 | - | - |
| Ralstoniasolanacearum | A6124 | S-26 | - | - |
| Ralstoniasolanacearum | A3292 | GMI-1000 | - | - |
Table 1: Bacterial strains used in this study and corresponding LAMP results.
Study design
DNA from Klebsiella spp. and Ralstonia solanacearum strains were tested individually, as well as in mixtures of two different genera or species. Individual and mixed DNA were further tested in the presence of plant tissue. Molecular tests of DNA included PCR followed by DGGE (denaturing gradient gel electrophoresis) analysis and LAMP. DNA from non-Klebsiella and non-Ralstonia bacteria were also tested with LAMP to determine specificity
Cultures of Klebsiella spp. and Ralstonia solanacearum strains were used individually, as well as in mixtures of two different genera or species, and tested with a colony-blot assay, in the absence and presence of plant tissue. Cultures were also inoculated into plants individually and as mixtures. Samples were taken from plants and tested with LAMP, DGGE, and colony-blot assay.
Plant inoculation
Plant inoculations were performed according to a previously established protocol [13,14]. Briefly, bacterial suspensions at 108 CFU/ ml were prepared from 48 hr cultures of test strains from TZC plates. Seeds of C. equisetifolia trees were collected from the University of Hawai'i at Mānoa campus, germinated on damp filter paper, placed in the dark and then transplanted to community pots. Established seedlings were transplanted to 6.7 cm pots containing Sunshine Mix #4 (Sun Gro Horticulture, Agawam, MA). Roots of 15-week-old seedlings were wounded by drawing a sterile scalpel through the soil at four sides of the pot in the root zone. Then, 10 ml of the bacterial suspension were pipetted into the soil after the wounding process. Older ironwood seedlings, ~17 weeks old, were inoculated with 50 ml of bacterial suspension in the same manner. Two plants were co-inoculated with Ralstonia solanacearum and Klebsiella variicola, but one was an older, 10-month old ironwood (Plant 1) that was used to try to produce bacterial ooze. The ironwood seedlings were then grown under the same conditions as described previously [14].
Plant sampling
Plant samplings were performed according to a previously established protocol [14]. Briefly, stem sections of plants were surface sterilized in 10% Clorox (final concentration: 0.8% sodium hypochlorite) for 30 seconds and triple-rinsed in sterile de-ionized water. Stem sections were then macerated in ~200 μl of sterile deionized water and the subsequent suspension was streaked onto TZC. Following a 24 h enrichment phase on TZC, the identity of the re-isolated bacteria was presumptively determined by appearance, and then confirmed using the various tests described hereafter. Tests were also performed directly on the extracted plant samples [15-19]. Bacterial ooze from plant samples was also collected by soaking stem sections in water to allow bacteria to diffuse from the plant material. For co-inoculated plants, five sample sections were taken from Plant 1, while eight were taken from a younger one (Plant 2), with sections being numbered sequentially away from the inoculum site. Tests were performed directly on bacterial ooze, as well as isolated colonies following a 12-h and 24-h enrichment.
DNA extraction
A Chelex DNA extraction was performed on isolated bacteria, as well as plant samples. Briefly, 0.75-1.0 ml of 40% Chelex 100 resin (Bio-Rad, Hercules, CA) in 1X TE buffer (10 mM Tris HCl and 1 mM EDTA at pH 8) with 10% Triton X-100 (Sigma-Aldrich, St. Louis, MO) was added to each sample. Samples were mixed by pipeting vigorously and vortexing and then heated to 95°C for 10 minutes on a digital heat block. Samples were stored at 4°C. Various other DNA extraction protocols were used on plant samples to remove inhibitors [20-24], including the PowerPlant Pro DNA isolation Kit (MO BIO Laboratories Inc., Carlsbad, CA).
DGGE
DGGE primers (341F(-GC)/907RA) used in this study were reported previously to amplify the variable V3 region of the 16S rRNA gene [25-27]. PCR reactions for 341F(-GC)/907RA were performed in a 25 μl reaction volume containing 1 μl bacterial DNA and 24 μl PCR reaction master mix [1 μl of each primer (10 μM), 0.13 μl GoTaq DNA polymerase (5 U/ μl) (Promega, Madison, WI), 5 μl 5X GoTaq Reaction Buffer (Promega, Madison, WI), 0.5 μl dNTP mix (10 mM) (Promega, Madison, WI) and 16.37 μl DNase/RNase free water]. PCR reaction conditions were as follows: an initial denaturing at 94°C for 5 min, followed by 25 cycles of denaturing at 94°C for 30 s, annealing at 57°C for 1.5 min, and elongation at 72°C for 1 min, with a final elongation at 72°C for 10 min. PCR products were resolved using DGGE. PCR primer sequences are listed in (Table 2).
| \OligonucleotidePrimerSequence(5’-3’) | Source/Reference | |
|---|---|---|
| LAMP | ||
| Hcp1-F3 | CCCCATACCTTTACAAGGCC | ThisStudy |
| Hcp1-B3 | CGTGGATCCTCCAGGTGATT | ThisStudy |
| Hcp1-FIP | GCTGAGTTTCACTCTTGAATACCAGCATTATGAACTTCGAACGT | ThisStudy |
| Hcp1-BIP | ACCGGGGTTAACTGTGGTGTACTGCAGGCTAACGCTTTCAAC | This Study |
| Hcp1-Loop | TGCAAACTTTCCTCGAATGACA | This Study |
| Hcp1-LoopProbe | /56-FAM/ACGCTGAGGACCCGGA TGCGAATGCGGATGCGGATGCCGATGCAAACTTTCCTCGAATGACA |
This Study |
| UGH-F3 | TGCCAGCAGACATTGACG | This Study |
| UGH-B3 | CCGACCACTACGAACGGT | This Study |
| UGH-FIP | CGATTGCGATCTGCGGCCTGCGCCAGTTTCTGGTAACCG | This Study |
| UGH-BIP | TCTGCAGCCAGCAGCAGATCGCCTGCGAGTATTTCTTCCG | This Study |
| UGH-Loop | GCGCATAAGCACTTTCCGG | This Study |
| UGH-LoopProbe | /56-FAM/ACGCTGAGGACCCGGATGCGAATGCG GATGCGGATGCCGATTTTGCGCATAAGCACTTTCCGG |
This Study |
| Quencherprobe | TCGGCATCCGCATCCGCATTCGCATCCGGGTCCTCAGCGT/3BHQ_1/ | Kubota et al., [11] |
| PCR | ||
| gyrA-A | CGCGTACTATACGCCATGAACGTA | Brisse and Verhoef, [42] |
| gyrA-C | ACCGTTGATCACTTCGGTCAGG | BrisseandVerhoef,[42] |
| DGGE | ||
| 341F | CCTACGGGAGGCAGCAG | Muyzeretal.,[25] |
| 341F(-GC) | (CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-)CCTACGGGAGGCAGCAG | Muyzeretal.,[25];Meyerand Kuever,2008 |
| 907RA | CCGTCAATTCATTTGAGTTT | Muyzeretal.,[26];Ishiiand Fukui,[27] |
/56-FAM/: 5’ 6- carboxyfluorescein; /3BHQ_1/: 3’ Black Hole Quencher 1
Table 2: Oligonucleotide primers used in this study*.
DGGE analysis was performed with a DGGE-2000 system (C.B.S Scientific Co.), using a 0.75 mm-thick 6% polyacrylamide gel (ratio of acrylamide to bis-acrylamide, 37.5:1) with a linear denaturing-agent gradient of 35-65% (100% denaturant agent was defined as 7 M urea with 40% formamide) that was submerged in 0.5X TAE buffer (40 mM Tris, 40 mM acetic acid, 1 mM EDTA; pH 7.4) at 60°C. PCR samples were mixed with 6 μl of dye solution [0.1% bromphenol blue (w/v), 70% glycerol (v/v)] and applied to the gels. Electrophoresis conditions were 5 h 30 min at 75 V. Gels were stained for 30 min in 1X TAE buffer with ethidium bromide and visualized using the Foto/Analyst Express System (Fotodyne Inc., Hartland, WI). Products were excised from gels and resuspended in sterile water [28]. Products were reamplified using 341F/907RA primers (Table 2), under the same reaction conditions, and then sequenced for identification. Sequences were queried in the National Center for Biotechnology Information (NCBI) database using the Basic Alignment Search Tool (BLAST) [29].
DNA sequencing
PCR products were cleaned for sequencing using ExoSAP-IT (Affymetrix, Santa Clara, CA), according to the manufacturer’s instructions. Cleaned PCR products were sequenced at the Greenwood Molecular Biology Facility at the University of Hawaiʻi sequencing facility, using each forward and reverse primer, according to specifications.
Colony-blot immunoassay
The colony blot immunoassay was performed according to methods established previously [30,31], with modifications. Briefly, 1 μl of bacterial suspension (ODA600=0.1), or plant extract, were spotted onto 0.45 μm nitrocellulose membranes (NitroBind, Osmonics, Inc., Minnetonka, MN) and allowed to dry completely. Dried nitrocellulose membranes were blocked in 5% PBS-milk for 1 h and then boiled in PBS for 10 min to eliminate endogenous enzymatic activity. Membranes were then washed three to five times using 0.05% PBS-Tween 20. Membranes were subsequently incubated with Klebsiella species antibody (73/28) (ThermoFisher Scientific, Waltham, MA) at a 1:2000 dilution for 30 min, followed by washing three times for five minutes each with 0.5% PBS-Tween 20. Membranes were then incubated with goat anti-mouse antibody conjugated to alkaline phosphatase (1:2000) (Southern Biotech, Birmingham, AL) for 30 min and washed three times for five minutes with 0.5% PBS-Tween 20. Membranes were developed with 1-Step NBT/BCIP (ThermoFisher Scientific, Waltham, MA) for 15-30 min, rinsed with distilled water, and air-dried. Colony immunoblots were analyzed under a dissecting microscope.
LAMP
LAMP primers developed in this study are shown in Table 2. Individual LAMP reactions were performed in triplicate and contained 5 μl sample, 5 μl primer master mix [F3 (0.2 μM), B3 (0.2 μM), FIP (1.6 μM), BIP (1.6 μM), Loop (0.8 μM), Loop Probe (0.08 μM), and Quencher probe (0.16 μM)] and 15 μl ISO-001nd Isothermal Mastermix (OptiGene, West Sussex, UK). Hcp1 and UGH primers were multiplexed to produce a reaction to detect both Klebsiella variicola and K. oxytoca, requiring 0.4 μM Quencher probe. Negative controls used 5 μl ddH2O and/or pathogen-free plant samples. LAMP reactions were run and analyzed using the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and a hand-held realtime assessment device (SMART-DART) (DiaGenetix Inc., Honolulu, HI), under the following conditions: 65°C for 30 min, with fluorescence readings being taken at 30-s or 1-min intervals. One drop of mineral oil was added to the tops of each sample when using the SMART-DART to prevent evaporation.
Sensitivity assay
Bacterial DNA was isolated using the Wizard Genome DNA Purification Kit (Promega, Madison, WI). DNA was quantified using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA) and diluted to a starting concentration of 1 ng/μl, with subsequent 10-fold serial dilutions made to reach a final concentration of 1 fg/μl. LAMP reactions were performed in triplicate, using ddH2O as the negative control.
Results
DGGE
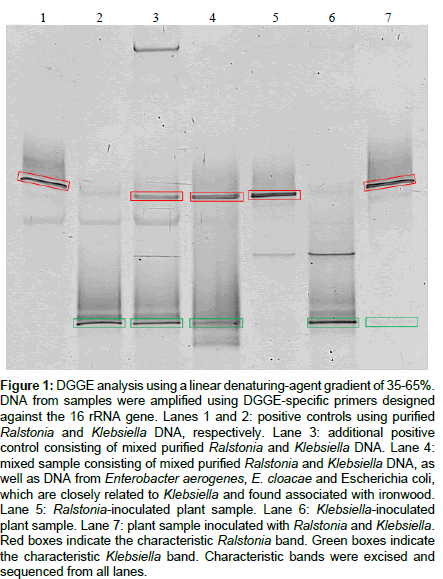
DGGE analyses were performed to determine its effectiveness in identifying Ralstonia and Klebsiella from inoculated ironwood. Samples were analyzed alongside positive controls (Figure 1). Positive controls consisting of purified DNA samples of Ralstonia, Klebsiella, and mixes of the two former (Figure 1, Lanes 1-3), show a characteristic Ralstonia and Klebsiella band (Figure 1, Lane 1 and Lane 2, respectively), with both bands appearing in the mixed sample (Figure 1, Lane 3). A mixed sample containing DNA from purified Ralstonia and Klebsiella, as well as DNA from Enterobacter aerogenes, E. cloacae and Escherichia coli, which are closely related to Klebsiella and found associated with ironwood, produced the characteristic Ralstonia and Klebsiella bands, however additional bands reflecting all of the additional bacteria contained within the sample were not present (Figure 1, Lane 4). Plants samples from ironwood inoculated with Ralstonia, Klebsiella, and both Ralstonia and Klebsiella produced the appropriate characteristic Ralstonia and Klebsiella bands (Figure 1, Lanes 5-7), with the Klebsiella band being faint in the plant sample with mixed infection (Figure 1, Lane 7). All characteristic bands were excised, re-amplified and sequenced. BLAST analysis of returned sequence data revealed that the characteristic Ralstonia and Klebsiella bands were indicative of each respective bacterium, thus demonstrating the utility of DGGE, and its potential ability to be used without sequencing by including appropriate controls. However, in the mixed sample, which contained other enteric bacteria (Figure 1, Lane 4), sequence data could not be obtained for the Klebsiella band. A strong band was produced for Ralstonia solanacearum, a weak band for Klebsiella, and no distinct bands for all other enteric bacteria. Similarities between bands of the enteric bacteria and Klebsiella could explain the lack of additional bands. Additionally, the Klebsiella band in the mixed plant sample was very faint (Lane 7). Inefficient recovery of DNA from the excised band is responsible for the inability to obtain sequence data.
Figure 1: DGGE analysis using a linear denaturing-agent gradient of 35-65%. DNA from samples were amplified using DGGE-specific primers designed against the 16 rRNA gene. Lanes 1 and 2: positive controls using purified Ralstonia and Klebsiella DNA, respectively. Lane 3: additional positive control consisting of mixed purified Ralstonia and Klebsiella DNA. Lane 4: mixed sample consisting of mixed purified Ralstonia and Klebsiella DNA, as well as DNA from Enterobacter aerogenes, E. cloacae and Escherichia coli, which are closely related to Klebsiella and found associated with ironwood. Lane 5: Ralstonia-inoculated plant sample. Lane 6: Klebsiella-inoculated plant sample. Lane 7: plant sample inoculated with Ralstonia and Klebsiella. Red boxes indicate the characteristic Ralstonia band. Green boxes indicate the characteristic Klebsiella band. Characteristic bands were excised and sequenced from all lanes.
Colony-blot assay
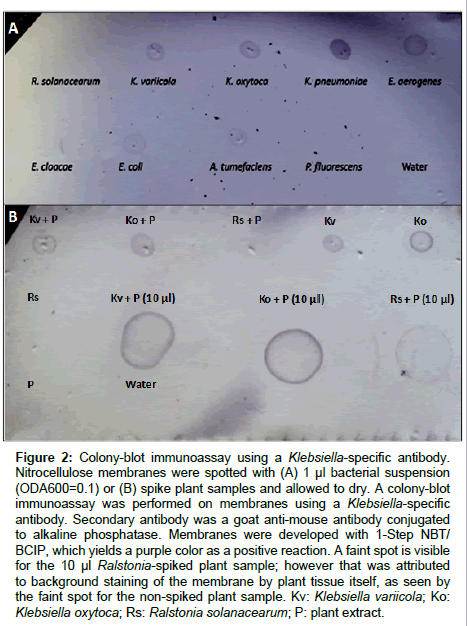
A Klebsiella-specific antibody was tested for specificity using pure bacterial suspensions in a colony-blot immunoassay format (Figure 2a). Positive reactions were obtained with all three Klebsiella spp. tested and negative reactions with R. solanacearum, P. fluorescens and the water control. However, Enterobacter aerogenes, E. coli, and Agrobacterium tumefasciens gave cross-reactions with the Klebsiella antibody assay. The colony-blot assay detected Klebsiella in spiked plant samples (Figure 2b). Positive controls made with pure bacterial suspensions of Klebsiella were positive, while negative controls consisting of Ralstonia, only macerated plant tissue, and water, were all negative. Plant samples containing Klebsiella variicola and K. oxytoca showed positive reactions, while plant samples containing Ralstonia did not, thus demonstrating potential for use in detecting Klebsiella from plant tissue samples.
Figure 2: Colony-blot immunoassay using a Klebsiella-specific antibody. Nitrocellulose membranes were spotted with (A) 1 μl bacterial suspension (ODA600=0.1) or (B) spike plant samples and allowed to dry. A colony-blot immunoassay was performed on membranes using a Klebsiella-specific antibody. Secondary antibody was a goat anti-mouse antibody conjugated to alkaline phosphatase. Membranes were developed with 1-Step NBT/ BCIP, which yields a purple color as a positive reaction. A faint spot is visible for the 10 μl Ralstonia-spiked plant sample; however that was attributed to background staining of the membrane by plant tissue itself, as seen by the faint spot for the non-spiked plant sample. Kv: Klebsiella variicola; Ko: Klebsiella oxytoca; Rs: Ralstonia solanacearum; P: plant extract.
LAMP
Two sets of LAMP primers were designed to specifically detect K. oxytoca and K. variicola. Both LAMP reactions were highly specific, only detecting their respective Klebsiella species and producing no reaction with any of the other bacteria tested in this study (Table 1), as well as negative controls. Positive LAMP reactions were recorded visually via turbidity, using the specified primers and a LAMP reaction master mix (dNTP mix (1.2 mM), 10X ThermoPol Reaction Buffer (New England Biolabs, Ipswich, MA) (2 mM), betaine (1 M), MgSO4 (4 mM), Bst DNA polymerase (New England Biolabs, Ipswich, MA) (8 U) and ddH2O) that was described previously [32,33]. Under the current protocol, positive reactions were visible in real-time and commonly occurred within 10-20 minutes
The sensitivity of the Hcp1-LAMP reaction was determined and DNA in the range of 5 ng to 5 pg showed positive reactions in approximately 10 minutes, while DNA in the range of 500 fg to 5 fg showed positive reactions in approximately 15 minutes. However, at the 5 fg range of DNA, positive reactions were not always obtained due to the low amount of DNA in the starting sample. The sensitivity of the UGH-LAMP reaction was less than that of the Hcp1-LAMP, with a detection limit of ~5 pg of DNA.
LAMP was performed on plant sample extracts following a 12-h and 24-h enrichment phase on solid media. All samples from known inoculated plants were positive by LAMP, while negative control plants were negative by LAMP. Non-Klebsiella bacteria that were present in the plant samples were tested separately and were negative with LAMP. In order to make a single test for both K. variicola and K. oxytoca, the LAMP primers were multiplexed into a single reaction. The multiplexed test was able to detect both Klebsiella spp.; however the sensitivity of the combined assay was compromised, as known to occur during multiplexing [34,35].
LAMP was further tested directly on plant sample extracts from two co-inoculated plants. Extracts were streaked onto plates and the first two sections of Plant 1 showed growth of Klebsiella and Ralstonia, while only Ralstonia grew from sections 3-5. In Plant 2, Klebsiella grew only from the first five sections. Initial testing using turbidity revealed that plant inhibitors did affect the LAMP reaction to a certain degree (data not shown). For real-time LAMP detection, extracts were processed, using various methods, and tests revealed no significant difference in results between direct boiling, 1:1 sample and TE buffer with boil, Chelex extraction, or GES buffer, using buffers 1 and 2 and sample at a 0.5:0.5:1 ratio. All Klebsiella-negative samples, including negative and Ralstonia controls, were all negative by LAMP. The LAMP reaction did not react with boiled samples from Plant 2 at sections 1, 3 and 4, TEbuffered samples from Plant 2 at section 2, and GES-buffered samples from Plant 1 at section 2. All other samples were positive by LAMP, including all Chelex-extracted samples. Included in the LAMP test was one Klebsiella-inoculated plant sample that had DNA extracted using the PowerPlant Pro DNA isolation kit, which is explicitly designed to extract DNA from plants and remove plant inhibitors. This sample was positive by LAMP, and reactions were the most robust; however, this kit is high in cost and therefore not practical for field use/studies. The SMART-DART hand-held real-time fluorescence monitoring device performed equally to the in-lab real-time machine.
Discussion
The genus Klebsiella is comprised of opportunistic pathogens that are frequently isolated from humans and animals [36]. In humans, Klebsiella pneumoniae and K. oxytoca are particularly involved with nosocomial infections [36,37], such as septicemia, pneumonia and urinary tract infection. In animals, Klebsiella spp. are mostly associated with sepsis, infections of the urinary and respiratory tracts, and mastitis, with Klebsiella pneumoniae and K. oxytoca being isolated frequently from domestic livestock [38-40]. These disease syndromes can cause serious economic consequences in some cattle herds [41]. The ecological habitats of Klebsiella include surface water, sewage, soils and plants, as well as mucosal surfaces of mammals [42]. Recently, a new species of Klebsiella was proposed, K. variicola, having clinical and plant-associated isolates [43].
In Guam, the association found between ironwood decline and Ralstonia led to the discovery of an association between Klebsiella and the same disease [11,12]. An association was also found between wetwood symptoms observed in infected trees and Klebsiella spp., which were determined to be Klebsiella oxytoca and K. variicola [13,14]. The current study, which focused on evaluation and design of diagnostic tools, will enable a more extensive survey of Guam ironwood trees to further investigate the association between Klebsiella- and Ralstoniainfected ironwood trees. DGGE, an immunoassay and a molecularbased assay (LAMP) were all tested for their ability to identify Klebsiella in ironwood.
DGGE is a method used to discern bacterial populations in mixed environmental samples. However, the complexity of DGGE makes it less desirable as a routine test for field studies. Additionally, results generally require DNA sequencing of bands following gel separation, which is not readily available to researchers in the field. Here, we attempted to circumvent the need for sequencing. Knowing what specific bacteria to look for and by running appropriate controls, we thought characteristic bands could be found that would be representative of Ralstonia and Klebsiella. While we were able to demonstrate this, the inability of DGGE to separate bands from closely related Enterobacter species contained in a mixed sample showed the possibility for false positive results. This fact, in combination with the laboriousness of DGGE, resulted in not recommending it as a possible test to survey Guam ironwood for Klebsiella infection.
Immunostrips (Agdia, Inc.) are available for detection of Ralstonia, but there are none for detection of Klebsiella. We explored the possibility of making an immunoassay that would be later commissioned into immunostrips for rapid detection of Klebsiella. We decided to test a commercial Klebsiella antibody for its potential as a rapid detection system, using a simple colony-blot test. As with most immunoassays, we observed cross-reactions with closely related bacteria. The Klebsiella antibody was made using Klebsiella aerogenes. Klebsiella species are constantly being reclassified into other genera and/or species. Interestingly enough, a very strong cross reaction was observed with Enterobacter aerogenes, even stronger than the interaction with Klebsiella oxytoca and K. variicola. Since this Klebsiella antibody reacted with a closely related Enterobacter sp. associated with ironwoods, this immunoassay is not recommended for routine field testing of Klebsiella-infected ironwood in Guam.
The potential for false positive results using the immunoassay and the inability to clearly separate bands using DGGE led to the design of a specific molecular-based assay using loop-mediated amplification technology. The LAMP assay proved to be the most useful in this study. It was able to specifically detect Klebsiella, having no positive reactions with non-Klebsiella bacteria, and the two primer sets allowed differential detection of Klebsiella oxytoca and K. variicola. LAMP primers in this study were designed to amplify a hemolysin-coregultaed protein (Hcp1) family type VI secretion system (T6SS) effector (K. variicola specific) and an unsaturated glucuronyl hydrolase (UGH) (K. oxytoca specific). These regions were found by whole-genome comparisons. BLAST searches of these genes resulted in matches only to each designated Klebsiella species, demonstrating that these primer sets should be highly specific for each Klebsiella species to which they were designed. LAMP tests using non-target bacteria confirmed the specificity of these genes as targets. Testing a broader panel of nontarget bacteria would improve specificity claims; however, in silico data from BLAST searches combined with test results provided in this study, together, suggest a high specificity.
As with any molecular-based assay, plant inhibitors are always a concern. Once an effective assay is developed, sample processing remains the underlying factor. The LAMP assays presented here were affected by plant inhibitors to an extent. Plant samples produced some false negatives, but there were no false positive results and Chelex extraction was able to remove enough inhibitors to allow proper detection directly from woody tissues. Ultimately, to avoid any problems with plant inhibitors, it is recommended to simply enrich samples on solid media [17,44,45]. Klebsiella grows extremely fast and a simple 12-h incubation on media will eliminate all false results.
LAMP was chosen because it is the most widely researched isothermal nucleic acid method, which allows greater trouble-shooting during development [46]. LAMP is comparable to equivalent PCR-, immunoassay- or culture-based detection methods [47], with detection limits as low as five copies [48]. Moreover, its isothermal nature makes it suitable for field diagnostics.
Molecular methods such as PCR have been evaluated for rapid identification of Klebsiella in human clinical specimens [37,49]. However, Klebsiella species are difficult to identify and are often misclassified in clinical microbiology laboratories [16,50,51]. Phenotypic distinction between K. pneumoniae and K. oxytoca isolates based on existing biochemical tests are time consuming, laborious and not very reliable [52]. Current procedures, such as BIOLOG and API systems, often fail to differentiate between species of Klebsiella [50,53]. Differentiation of Klebsiella is a key component of clinical responses to infection. In Japan, in 2013, a patient with sepsis caused by Klebsiella variicola died due to misidentification as Klebsiella pneumoniae by a commonly used automated identification system [54]. The need for diagnostic tests that can properly identify Klebsiella at the species level has been nationally recognized, as the National Institute of Health and the National Institute of Allergy and Infectious Disease recently put out a request for proposal for an R01 grant that included Klebsiella as a target organism (Funding Opportunity Announcement Number RFAAI- 14-019).
The LAMP assays developed for Klebsiella have potential as clinical diagnostic tests because LAMP is less sensitive than PCR to inhibitory substances present in biological samples [55] such as serum [56], CSF [57], swabs [58], and heat-treated blood [59], saving time and money required for sample processing steps [60]. As a result, this technology has been used widely for molecular detection of several microorganisms, including clinical and plant-associated bacterial [32,61-73], fungal [74-76], viral [47,77-84], and parasitic [57,59,85-88] pathogens, and is therefore suitable as a rapid field test
Due to the isothermal nature of LAMP, it offers an easy and rapid assay for clinical medicine [89]. The LAMP reactions presented in this study not only specifically detect Klebsiella, but also distinguish K. oxytoca and K. variicola. Moreover, the target regions that these LAMP primers were designed to could become the new focus for developing Klebsiella-detection systems, having application to numerous molecular detection platforms. In conjunction with portable real-time fluorescence monitoring devices, such as the SMART-DART, these LAMP reactions could provide beneficial point-of-care diagnostics for clinical, veterinary and agricultural Klebsiella spp.
Acknowledgements
This work was supported in part by USDA National Institute for Food and Agriculture, Project HAW00987-H, administered by the College of Tropical Agriculture and Human Resources, University of Hawaiʻi at Mānoa. This work was also supported in part by WPDN-201303063-01 and by projects administered by the University of Guam: US Forest Service project 12-DG-11052021-236, USDA NIFA, RREA project 231395 and USDA NIFA, McIntire-Stennis project 1005476. The authors thank Daniel Jenkins and Ryo Kubota of DiaGenetix Inc. (Honolulu, HI) for providing the Isothermal Mastermix and SMART-DART
References
- Morton JF (1980) The Australian pine or beefwood (Casuarinaequisetifolia L.), an invasive "weed" tree in Florida. ProcFla State HortSoc 93: 87-95.
- Schlub KA (2010) Investigating the ironwood tree (Casuarinaequisetifolia) decline in Guam using applied multinomial modeling: Louisiana State University.
- Mersha Z, Schlub RL, Spaine PO, Smith JA, Nelson SC (2010) Visual and quantitative characterization of ironwood tree (Casuarinaequisetifolia) decline on Guam. Phytopathology 100: S82
- Mersha Z, Schlub RL, Moore L (2009) The state of ironwood (Casuarinaequisetifolia subsp. equisetifolia) decline of the Pacific island of Guam. Phytopathology 99: S85.
- Schlub RL, Schlub KA, Alvarez AM, Aime CM, Cannon PG, et al. (2012) Integrated perspective on tree decline of ironwood (Casuarinaequisetifolia) on Guam. Proceedings of the 60th Annual Western International Forest Disease Work Conference: October 8-12, 2012 2012; Lake Tahoe, CA: 51-60.
- Schlub RL, Kubota R, Alvarez AM (2013) Casuarinaequisetifolia decline in Guam linked to colonization of woody tissues by bacteria. Phytopathology 103: S2.128.
- Schlub RL, Mendi R, Aiseam C, Mendi R, Davis J, et al. (2012) Survey of wood decay fungi on Casuarinaequisetifolia (ironwood) on the islands of Guam and Saipan. Phytopathology 102: S416.
- Orian G (1961) Diseases of Filao (Casuarinaequisetifolia) forest in Mauritius. Revue agricoleetsucrière de l'île Maurice 40: 17-45.
- Ali MIM, Anuratha CS, Sharma JK (1991) Bacterial wilt of Casuarinaequisetifolia in India. Eur J Forest Pathol 21: 234-238.
- Schlub RL, Moore A, Marx BD, Schlub KA, Kennaway L, et al. (2011) Decline of Casuarinaequisetifolia (ironwood) trees on Guam: symptomology and explanatory variables. Phytopathology 101: S216.
- Kubota R, Schell M, Peckham G, Rue J, Alvarez AM, et al. (2011) In silico genomic subtraction guides development of highly accurate, DNA-based diagnostics for Ralstoniasolanacearum race 3 biovar 2 and blood disease bacterium. J Gen Plant Pathol 77: 182-193.
- Kubota R, Vine BG, Alvarez AM, Jenkins DM (2008) Detection of Ralstoniasolanacearum by loop-mediated isothermal amplification. Phytopathology 98: 1045-1051.
- Ayin CM, Schlub RL, Alvarez AM (2013) Identification of bacteria associaed with decline of ironwood trees (Casuarinaequisetifolia) in Guam. Phytopathology 103: S2, 10.
- Ayin CM, Schlub RL, Yasuhara-Bell J, Alvarez AM (2015) Identification and characterization of bacteria associated with decline of ironwood (Casuarinaequisetifolia) in Guam. Australas Plant Pathol 44: 225-234.
- Schneider KL, Marrero G, Alvarez AM, Presting GG (2011) Classification of plant associated bacteria using RIF, a computationally derived DNA marker. PLoS One 6: e18496.
- Alves MS, Dias RC, de Castro AC, Riley LW, Moreira BM (2006) Identification of clinical isolates of indole-positive and indole-negative Klebsiella spp. J ClinMicrobiol 44: 3640-3646.
- Schaad NW, Jones JB, Chun W (2001) Laboratory Guide for Identification of Plant Pathogenic Bacteria, APS Press, St. Paul, Minnesota, USA.
- Murdoch CW, Campana RJ (1983) Bacterial species associated with wetwood of elm. Phytopathology 73: 1270-1273.
- Tiedemann G, Bauch J, Bock E (1977) Occurrence and significance of bacteria in living trees of Populusnigra L. Eur J Forest Pathol 7: 364-374.
- Bellstedt DU, Pirie MD, Visser JC, de Villiers MJ, Gehrke B (2010) A rapid and inexpensive method for the direct PCR amplification of DNA from plants. Am J Bot 97: e65-68.
- De Boer SH, Ward LJ, Li X, Chittaranjan S (1995) Attenuation of PCR inhibition in the presence of plant compounds by addition of BLOTTO. Nucleic Acids Res 23: 2567-2568.
- Llop P, Caruso P, Cubero J, Morente C, López MM (1999) A simple extraction procedure for efficient routine detection of pathogenic bacteria in plant material by polymerase chain reaction. J Microbiol Methods 37: 23-31.
- Singh RP, Nie X, Singh M, Coffin R, Duplessis P (2002) Sodium sulphite inhibition of potato and cherry polyphenolics in nucleic acid extraction for virus detection by RT-PCR. J Virol Methods 99: 123-131.
- Samarakoon T, Wang SY, Alford MH (2013) Enhancing PCR amplification of DNA from recalcitrant plant specimens using a trehalose-based additive. Appl Plant Sci 1.
- Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695-700.
- Muyzer G, Teske A, Wirsen CO, Jannasch HW (1995) Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol 164: 165-172.
- Ishii K, Fukui M (2001) Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microbiol 67: 3753-3755.
- Díez B, Pedrós-Alió C, Marsh TL, Massana R (2001) Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl Environ Microbiol 67: 2942-2951.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J MolBiol 215: 403-410.
- Alvarez AM, Kaneshiro WS, Vine BG (2005) Diversity of Clavibactermichiganensis subsp. michiganensis populations in tomato seed: What is the significance? ActaHort 695: 205-213.
- Kaneshiro WS: Detection and characterization of virulent, hypovirulent, and nonvirulentClavibactermichiganensis subsp. michiganensis. Honolulu: University of Hawai`i at Manoa; 2003.
- Kubota R, Vine BG, Alvarez AM, Jenkins DM (2008) Detection of Ralstoniasolanacearum by loop-mediated isothermal amplification. Phytopathology 98: 1045-1051.
- Yasuhara-Bell J, Kubota R, Jenkins DM, Alvarez AM (2013) Loop-mediated amplification of the Clavibactermichiganensis subsp. michiganensismicA gene is highly specific. Phytopathology 103: 1220-1226.
- Kanagawa T (2003) Bias and artifacts in multitemplate polymerase chain reactions (PCR). J BiosciBioeng 96: 317-323.
- Polz MF, Cavanaugh CM (1998) Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol 64: 3724-3730.
- Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. ClinMicrobiol Rev 11: 589-603.
- Neuberger A, Oren I, Sprecher H (2008) Clinical impact of a PCR assay for rapid identification of Klebsiella pneumoniae in blood cultures. J ClinMicrobiol 46: 377-379.
- Brisse S, DuijkerenEv (2005) Identification and antimicrobial susceptibility of 100 Klebsiella animal clinical isolates. Vet Microbiol 105: 307-312.
- Kikuchi N, Kagota C, Nomura T, Hiramune T, Takahashi T, et al. (1995) Plasmid profiles of Klebsiella pneumoniae isolated from bovine mastitis. Vet Microbiol 47: 9-15.
- Wilson DJ, Gonzalez RN, Das HH (1997) Bovine mastitis pathogens in New York and Pennsylvania: prevalence and effects on somatic cell count and milk production. J Dairy Sci 80: 2592-2598.
- Waller KP, Unnerstad H (2004) Klebsiella mastitis: a potential threat to milk production. SvenskVeterinärtidning 56: 11-17.
- Brisse S, Verhoef J (2001) Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J SystEvolMicrobiol 51: 915-924.
- Rosenblueth M, Martínez L, Silva J, Martínez-Romero E (2004) Klebsiella variicola, a novel species with clinical and plant associated isolates. SysetmApplMicrobiol 27: 27-35.
- Brisse S, Grimont F, Grimont PAD: The genus Klebsiella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes. vol. 6. New York: Springer; 2006: 159-196.
- Gephardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, et al. 1981 Manual of Methods for General Bacteriology, ASM Press, Washington D.C.
- Craw P, Balachandran W (2012) Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip 12: 2469-2486.
- Fujino M, Yoshida N, Yamaguchi S, Hosaka N, Ota Y, et al. (2005) A simple method for the detection of measles virus genome by loop-mediated isothermal amplification (LAMP). J Med Virol 76: 406-413.
- Iwamoto T, Sonobe T, Hayashi K (2003) Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J ClinMicrobiol 41: 2616-2622.
- Kovtunovych G, Lytvynenko T, Negrutska V, Lar O, Brisse S, et al. (2003) Identification of Klebsiella oxytoca using a specific PCR assay targeting the polygalacturonasepehX gene. Res Microbiol 154: 587-592.
- Monnet D, Freney J (1994) Method for differentiating Klebsiella planticola and Klebsiella terrigena from other Klebsiella species. J ClinMicrobiol 32: 1121-1122.
- Hansen DS, Aucken HM, Abiola T, Podschun R (2004) Recommended test panel for differentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. J ClinMicrobiol 42: 3665-3669.
- Chander Y, Ramakrishnan MA, Jindal N, Hansen K, Goyal SM (2011) Differentiation of Klebsiella pneumoniae and K. oxytoca by multiplex polymerase chain reaction. Intern J Appl Res Vet Med 9: 138-142.
- Monnet D, Freney J, Brun Y, Boeufgras JM, Fleurette J (1991) Difficulties in identifying Klebsiella strains of clinical origin. ZentralblBakteriol 274: 456-464.
- Seki M, Gotoh K, Nakamura S, Akeda Y, Yoshii T, et al. (2013) Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. J Med Microbiol 62: 801-803.
- Kaneko H, Kawana T, Fukushima E, Suzutani T (2007) Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J BiochemBiophys Methods 70: 499-501.
- Ihira M, Sugiyama H, Enomoto Y, Higashimoto Y, Sugata K, et al. (2010) Direct detection of human herpesvirus 6 DNA in serum by variant specific loop-mediated isothermal amplification in hematopoietic stem cell transplant recipients. J Virol Methods 167: 103-106.
- Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, et al. (2008) African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol 38: 589-599.
- Enomoto Y, Yoshikawa T, Ihira M, Akimoto S, Miyake F, et al. (2005) Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. J ClinMicrobiol 43: 951-955.
- Poon LLM, Wong BWY, Ma EHT, Chan KH, Chow LMC, et al. (2006) Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. ClinChem 52: 303-306.
- Mori Y, Notomi T (2009) Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother 15: 62-69.
- Enosawa M, Kageyama S, Sawai K, Watanabe K, Notomi T, et al. (2003) Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis. J ClinMicrobiol 41: 4359-4365.
- Hanaki K, Sekiguchi J, Shimada K, Sato A, Watari H, et al. (2011) Loop-mediated isothermal amplification assays for identification of antiseptic- and methicillin-resistant Staphylococcus aureus. J Microbiol Methods 84: 251-254.
- Harper SJ, Ward LI, Clover GR (2010) Development of LAMP and real-time PCR methods for the rapid detection of Xylellafastidiosa for quarantine and field applications. Phytopathology 100: 1282-1288.
- Koide Y, Maeda H, Yamabe K, Naruishi K, Yamamoto T, et al. (2010) Rapid detection of mecA and spa by the loop-mediated isothermal amplification (LAMP) method. LettApplMicrobiol 50: 386-392.
- Kubota R, LaBarre P, Singleton J, Beddoe A, Weigl BH, et al. (2011) Non-Instrumented Nucleic Acid Amplification (NINA) for Rapid Detection of Ralstoniasolanacearum Race 3 Biovar 2. BiolEng Trans 4: 69-80.
- Lalande V, Barrault L, Wadel S, Eckert C, Petit JC, et al. (2011) Evaluation of a loop-mediated isothermal amplification assay for diagnosis of Clostridium difficile infections. J ClinMicrobiol 49: 2714-2716.
- Lin GZ, Zheng FY, Zhou JZ, Gong XW, Wang GH, et al. (2011) Loop-mediated isothermal amplification assay targeting the omp25 gene for rapid detection of Brucella spp. Mol Cell Probes 25: 126-129.
- McKenna JP, Fairley DJ, Shields MD, Cosby SL, Wyatt DE, et al. (2011) Development and clinical validation of a loop-mediated isothermal amplification method for the rapid detection of Neisseria meningitidis. DiagnMicrobiol Infect Dis 69: 137-144.
- Misawa Y, Yoshida A, Saito R, Yoshida H, Okuzumi K, et al. (2007) Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J Infect Chemother 13: 134-140.
- Neonakis IK, Spandidos DA, Petinaki E (2011) Use of loop-mediated isothermal amplification of DNA for the rapid detection of Mycobacterium tuberculosis in clinical specimens. Eur J ClinMicrobiol Infect Dis 30: 937-942.
- Temple TN, Johnson KB (2011) Evaluation of loop-mediated isothermal amplification for rapid detection of Erwiniaamylovora on pear and apple fruit flowers. Plant Dis 95: 423-430.
- Temple TN, Stockwell VO, Johnson KB (2007) Development of a rapid detection method for Erwiniaamylovora by loop-mediated isothermal amplification (LAMP). XI International Workshop on Fire Blight 739: 497-503.
- Yamazaki W, Seto K, Taguchi M, Ishibashi M, Inoue K (2008) Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification. BMC Microbiol 8: 94.
- Lucas S, da Luz Martins M, Flores O, Meyer W, Spencer-Martins I, et al. (2010) Differentiation of Cryptococcus neoformans varieties and Cryptococcus gattii using CAP59-based loop-mediated isothermal DNA amplification. ClinMicrobiol Infect 16: 711-714.
- Niessen L, Vogel RF (2010) Detection of Fusariumgraminearum DNA using a loop-mediated isothermal amplification (LAMP) assay. Int J Food Microbiol 140: 183-191.
- Sun J, Najafzadeh MJ, Vicente V, Xi L, de Hoog GS (2010) Rapid detection of pathogenic fungi using loop-mediated isothermal amplification, exemplified by Fonsecaea agents of chromoblastomycosis. J Microbiol Methods 80: 19-24.
- Curtis KA, Rudolph DL, Owen SM (2008) Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP). J Virol Methods 151: 264-270.
- Curtis KA, Rudolph DL, Owen SM (2009) Sequence-specific detection method for reverse transcription, loop-mediated isothermal amplification of HIV-1. J Med Virol 81: 966-972.
- Dinh DT, Le MT, Vuong CD, Hasebe F, Morita K (2011) An Updated Loop-Mediated Isothermal Amplification Method for Rapid Diagnosis of H5N1 Avian Influenza Viruses. Trop Med Health 39: 3-7.
- Hatano B, Goto M, Fukumoto H, Obara T, Maki T, et al. (2011) Mobile and accurate detection system for infection by the 2009 pandemic influenza A (H1N1) virus with a pocket-warmer reverse-transcriptase loop-mediated isothermal amplification. J Med Virol 83: 568-573.
- Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, et al. (2006) Development of H5-RT-LAMP (loop-mediated isothermal amplification) system for rapid diagnosis of H5 avian influenza virus infection. Vaccine 24: 6679-6682.
- Kurosaki Y, Grolla A, Fukuma A, Feldmann H, Yasuda J (2010) Development and evaluation of a simple assay for Marburg virus detection using a reverse transcription-loop-mediated isothermal amplification method. J ClinMicrobiol 48: 2330-2336.
- Okafuji T, Yoshida N, Fujino M, Motegi Y, Ihara T, et al. (2005) Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J ClinMicrobiol 43: 1625-1631.
- Parida M, Posadas G, Inoue S, Hasebe F, Morita K (2004) Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J ClinMicrobiol 42: 257-263.
- Bakheit MA, Torra D, Palomino LA, Thekisoe OM, Mbati PA, et al. (2008) Sensitive and specific detection of Cryptosporidium species in PCR-negative samples by loop-mediated isothermal DNA amplification and confirmation of generated LAMP products by sequencing. Vet Parasitol 158: 11-22.
- Lau YL, Meganathan P, Sonaimuthu P, Thiruvengadam G, Nissapatorn V, et al. (2010) Specific, sensitive, and rapid diagnosis of active toxoplasmosis by a loop-mediated isothermal amplification method using blood samples from patients. J ClinMicrobiol 48: 3698-3702.
- Matovu E, Kuepfer I, Boobo A, Kibona S, Burri C (2010) Comparative detection of trypanosomal DNA by loop-mediated isothermal amplification and PCR from flinders technology associates cards spotted with patient blood. J ClinMicrobiol 48: 2087-2090.
- Nkouawa A, Sako Y, Li T, Chen X, Wandra T, et al. (2010) Evaluation of a loop-mediated isothermal amplification method using fecal specimens for differential detection of Taenia species from humans. J ClinMicrobiol 48: 3350-3352.
- Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T (2001) Loop-mediated isothermal amplification reaction using a nondenatured template. ClinChem 47: 1742-1743.
Copyright: © 2015 Yasuhara-Bell J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.