Indexed In
- Open J Gate
- Academic Keys
- ResearchBible
- China National Knowledge Infrastructure (CNKI)
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- CABI full text
- Publons
- Geneva Foundation for Medical Education and Research
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Original Research Article - (2019) Volume 7, Issue 5
Seroreactivity of Populations Living in Endemic Area of Burkina Faso to Plasmodium falciparum Alpha-Helical Coiled Coil Proteins Motifs by Protein Microarray
Oumarou Ouédraogo1,2*, Luisa Nunziangeli3, Edith C. Bougouma1, Youssouf Kaboré1, Amidou Diarra1, Blami Koté4, Alfred B. Tiono1, Giampietro Corradin5, Valentina Mangano6, David Modiano6, Yves Traoré2, Sodiomon B. Sirima1, Roberta Spaccapelo7 and Issa Nébié12Department of Biochemistry and Microbiology, University of Ouagadougou, Ouagadougou, Burkina Faso
3Polo d'Innovazione di Genomica, Genetica e Biologia, Terni, Italy
4University of Ouahigouya, Ouahigouya, Burkina Faso
5Biochemistry Department, University of Lausanne, 1066 Epalinges, Switzerland
6Univesrita la Sapienza, Rome, Italy
7Department of Experimental Medicine, University of Perugia, Perugia, Italy
Received: 13-Jun-2019 Published: 28-Aug-2019, DOI: 10.35248/2329-891X.19.7.320
Abstract
The naturally acquired immunity is one of the models of immunity that is exploited for malaria antigens discovery. We have used multiplex protein microarrays of 92 P. falciparum alpha-helical coiled coil protein motifs to screen plasma samples obtained from 1113 children and adults belonging to three sympatric ethnic groups from malaria endemic area of Burkina Faso. We have investigated the influence of some factors such as age, ethnicity, hemoglobin genotype and sex on the antigens reactivity and the IgG antibody level. We also investigated specifically, the influence of the factors mentioned above on the 36 antigens with the highest antibody prevalence and intensity. As expected, the findings of the study confirmed the positive correlation between age, antigens reactivity and IgG antibody level. Except for three antigens (MSP2_3D7, MR260, and As182.20), the IgG level was higher in adults compared to children. The Fulani ethnic group recognized more antigens with the highest IgG values compared to the sympatric groups of Mossi and Rimaibé, except for six antigens (LR166, LR146, AS182.15, MR214, MR236A, and MSP3). In General, the hemoglobin type and sex did not have any influence on the IgG antibody reactivity and intensity. Except for four antigens (MR232, MR261A, MSP2_3D7, and NANP) 10), there was statistically no IgG intensity difference between people with normal hemoglobin genotype (AA) and the non-(AA) volunteers. Our study has demonstrated that protein microarrays of alpha-helical coiled-coil proteins motifs are suitable to screen the naturally acquired immunity. The findings of the study should be considered in any strategy for new antigen related to alpha-helical coiled coil protein segments discovery for a potential vaccine clinical development.
Keywords
Plasmodium falciparum; Protein microarray; Naturally acquired immunity; Malaria vaccine
Introduction
Malaria still remains a major public health problem worldwide. There were an estimated 219 million cases and 435,000 related deaths due to malaria in 2017 [1]. To date no licensed malaria vaccine is available. The increasing resistance of P. falciparum to the common antimalarial drugs and the resistance of the malaria vectors to usual insecticides is stressing the urgent need of an effective malaria vaccine [2,3]. The Naturally Acquired Immunity (NAI) developed in people naturally exposed to malaria parasites in the endemic area is one of the models of immunity which is exploited for malaria vaccine antigens discovery [4]. So far most of the seroepidemiological studies which aimed to discover malaria vaccines were focused on a limited number of well-known antigens of malaria parasites. They also employed traditional methods of screening such as ELISA that are laborintensive and time-consuming. The complete sequencing of the genome of the most virulent malaria parasite Plasmodium falciparum [5] and the availability of high throughput techniques such as protein microarray offers the ability to screen a large panel of malaria antigens and to faster the identification of potential target antigens that could undergo clinical development as potential malaria vaccines candidates. Several studies have used protein microarrays of recombinant proteins to explore the NAI with the aim to discover new malaria potential vaccines [6-10]. The high Adenine and Thymine (AT) composition of the Plasmodium falciparum genome makes the production of its recombinant proteins challenging. Moreover, the purification of Plasmodium recombinant proteins is also a limiting factor. They can also yield some folding and solubility problems. Moreover, these proteins are most of the time polymorphic.
Synthetic peptides with alpha helicoidal coiled-coil motifs have some interesting features such as the lack of folding problems, the ease of purification. They also don't have any solubility issues [11].
We, therefore, propose to use the high throughput protein microarrays of Plasmodium falciparum synthetic peptides to explore the naturally acquired immunity developed in people across ethnic groups and age range living in a malaria-endemic area.
Materials and Methods
Ethical statement
The samples used in our study are part of a seroepidemiological study which received approval from the Ethical committees of the Health ministry of Burkina Faso (deliberation N° 2007-48) and the University of Oxford. The study volunteers or their guardians gave written informed consent for participation.
Study site and samples collection
The study was carried out in 4 rural villages of Burkina Faso, Northeast (Barkoundouba and Barkoumbilen) and East (Bassy and Zanga) of Ouagadougou, the capital city.
The study site has been already described elsewhere [12]. Plasma samples were obtained from children and adults belonging to 3 sympatric ethnic groups (Mossi, Fulani, and Rimaïbé). For the negative controls, the plasma samples were obtained from Italian malaria-naive volunteers.
Hemoglobin genotyping
The method used for hemoglobin genotyping was already described elsewhere [12]. Briefly, genomic DNA was extracted from whole blood using the Nucleon BACC2 kit. The rs334 (A→HbA/T → HbS) and rs33930165 (G → HbA/A → HbC) SNPs at the HBB locus were genotyped using the Sequenom MassArray System [13].
Plasmodium falciparum antigens
We used microarrays containing in total 92 Plasmodium falciparum synthetic peptides of which 88 were a-helical coiled-coil segments and 4 well known synthetic malaria antigens (MSP3, MSP2-3D7, (NANP)10 and P27A) as positive controls (Supplementary Table 1), [11] and unpublished results). The bioinformatic screening and the chemical synthesis procedure are described elsewhere [11].
Protein microarray immunoassay
Manufacturing of the microarrays: For microarrays construction, the synthetic peptides were resuspended in PBS 1 X to a concentration of 300 μg/mL. Peptides were printed to aldehyde -derivatized glass microscope slides (CEL Associates, Houston, TX, USA) using computer-controlled high-speed robotics (Microgrid II Biorobotics®. Genomic Solutions Ltd. Huntingdon. Cambridgeshire. the UK). The solutions of antigens were transferred from 384-well microtiter plates (Greiner bio-one. the USA) onto glass slides by using stainless steel solid pins of 200 μm of diameter.
The microarrays consist of 16 × 16 matrices that included: (i) the P. falciparum antigens printed in two replicates; (ii) HSA as negative controls; (iii) An anti-mouse IgG and Human purified IgG as signal controls; (iv) PBS used as carry-over controls (blanks).
The printing of the peptides was performed inside the printer cabinet at a temperature (22°C-24°C) and humidity (45%-55%). Printed slides were kept overnight inside the printing cabinet before removal in order to optimize the peptides binding. Slides were subsequently kept in boxes at room temperature in the presence of silica gel bags.
Immunoassay: Printed slides were incubated for 1 hour at room temperature with a blocking buffer (PBS 1X, BSA 2%) in order to prevent no specific binding. A hydrophobic pen (ImmEdgeTM PEN. Vector Laboratories Inc., Burlingame, USA) was used to delimit each array prior to the blocking step in order to contain reagents and samples within arrays areas and prevent cross-contamination. After the incubation step, slides were washed with washing buffer (PBS 1X, Tween 20 at 0.01%) in order to remove unbound material.
Plasma samples were diluted at 1:100 in a dilution buffer (PBS 2X, BSA 2%, Tween 20 at 0.01 %) and 50 μl of the diluted sample were allowed to react with each area for 1 hour at room temperature. Slides were washed with washing buffer to remove no specific material. To reveal IgG bound to the printed peptides, the slides were incubated 20 minutes at room temperature with Alexa 647-labeled goat anti-human IgG secondary antibody (SouthernBiotech) at a concentration of 10 μg/mL. Unbound secondary antibody was removed by washing with the washing buffer. Then, the slides were dried by centrifugation at 3000 rpm for 1 minute prior to the detection of the fluorescence. All the plasma samples were tested in double.
Detection and quantification of array components fluorescence: The fluorescents signal from the slides were read at 633 nm by using a ScanArray Gx Scanner (Perkin-Elmer, Cambridge, UK). The fluorescent signal from each array component was visualized in a pseudocolor scale corresponding to increasing fluorescence (from dark-blue; blue; green; red to white).
Images generated are saved as JPEG and TIF files. JPEG files were used for signal visualization. TIF files were used for signal quantification with the ScanArrayExpressTM software (Perkin-Elmer).
Treatment of microarray data and statistical analysis
Quantified array signals were analyzed using Excel (Microsoft). Measurements for each spot were corrected against the internal PBS negative control to identify signals above background. Four replicate measurements were calculated for each antigen. Negative or Zero values after the background subtraction were assigned a net value of 0. The data were normalized before statistical analysis was performed.
To determine the positive reactivity cut-off of a sample to a given antigen. We calculated the mean from the 20 Italian negative controls. A sample was reactive if the intensity of the antigen-bounded IgG was above the upper limit of the confidence interval of the mean of the negative control.
The statistical analysis of the data was performed using Microsoft Excel and STATA software (Version 11.0, College Station, TX: StataCorp; 2011).
The cut-off value of a positive reactivity was defined for each antigen. A sample reactivity to a given peptide was positive when the reactivity is above the upper limit of the confidence interval of the mean of the Italian negative controls. A classical t-test of the hypothesis was used for means comparisons across distinct groups.
A p-value<0.05 was considered as the threshold for the statistical significance of hypothesis tests.
Results
Characteristics of study participants
Plasma samples were obtained during a cross-sectional study carried out in August 2007, from 1113 naturally exposed individuals to malaria in Burkina Faso. Samples from 20 Italians malaria naïve individuals served as negative controls. The characteristics of the study participants are described in Table 1.
| Sex | No. |
|---|---|
| Female (51.48) | 573 |
| Male (48.52) | 540 |
| Age (years) | |
| ≤ 5 (19.59) | 218 |
| 5-15 (41.69) | 464 |
| >15 (38.72) | 431 |
| Ethnicity | |
| Fulani (36.66) | 408 |
| Mossi (30.46) | 339 |
| Rimaibé (32.88) | 366 |
| Hemoglobin type | |
| AA (75.00) | 762 |
| AC (18.11) | 184 |
| AS (5. 91) | 60 |
| CC (0.30) | 3 |
| SC (0.69) | 7 |
Table 1: Characteristic of study participants.
Prevalence and intensity of the positive antibody reactivity among the study population
The prevalence of positive reaction to all the peptides and intensity of IgG response are shown in Supplementary Table 2.
Influence of age on antigens reactivity an antibody intensity
We found that the number of recognized peptides, as well as the level of IgG, was lower in children under 5 compared to the other age group. Then, we also investigated the total amount of total IgG to all of the recognized peptides in children and adults. As expected, the total amount of IgG was higher in adults compared to children.
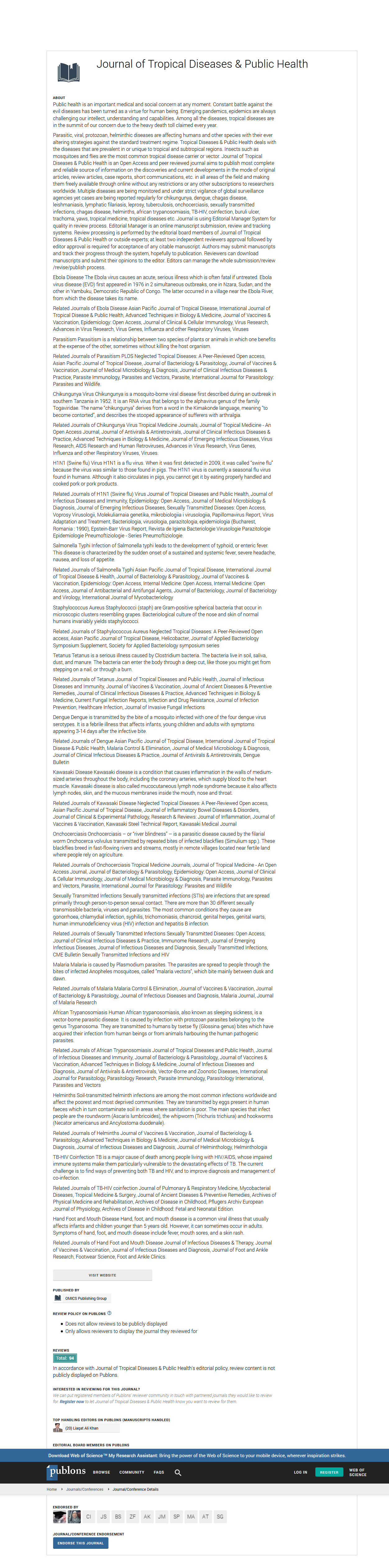
Figure 1 shows the breadth of recognized peptides and the intensity of IgG response in the different age groups.
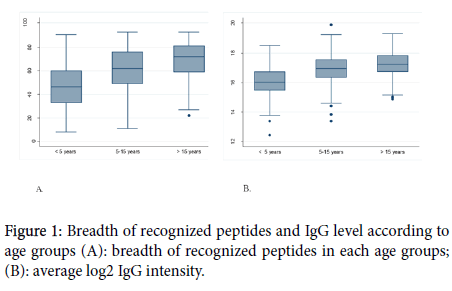
Figure 1. Breadth of recognized peptides and IgG level according to age groups (A): breadth of recognized peptides in each age groups; (B): average log2 IgG intensity.
The average number of recognized peptides in adults and children were respectively 65 and 59 and the difference between the two groups was statistically significant (p<0.0001). The same finding was observed concerning the level of IgG antibody with adults having the highest level of antibody (p<0.0001).
The peptides with a positive IgG prevalence greater than 50% and intensity ratio greater than 5 were selected. In total 36 antigens comprising the well-known antigens (MSP3, MSP2_3D7, (NANP)10 and 27A [14] were selected and the influence of age on the specific IgG antibody response to each of those antigens was investigated. There was statistically a significant difference between the IgG antibody response to 33 antigens between adults and children with a higher response in adults. The results are shown in Table 2.
| Name | Protein ID | Mean Intensity log IgG (95% CI) | ||
|---|---|---|---|---|
| Children ( ≤ 15 ans) | Adults ( >15 ans) | p-value | ||
| LR166 | PFA0635c | 5.92 (5.82-6.02) | 6.19 (6.08-6.30) | 0.0006 |
| LR146 | PFB0145c | 6.30 (6.14- 6.47) | 6.78 (6.61-6.95) | 0.0001 |
| LR149 | PFB0145c | 5.99 (5.86-6.12) | 6.44 (6.31-6.58) | 0 |
| LR150A | PFB0145c | 6.80 (6.64-6.96) | 7.22 (7.06-7.39) | 0.0003 |
| LR150B | PFB0145c | 6.65 (6.48-6.82) | 7.14 (6.97- 7.30) | 0.0001 |
| MR219 | PFB0315w | 6.05 (5.85-6.24) | 6.64 (6.42-6.85) | 0.0001 |
| MR220 | PFC0345w | 6.78 (6.64- 6.92) | 7.08 (6.92-7.23) | 0.0081 |
| LR157A | PFD0110w | 6.77 (6.62-6.93) | 7.03 (6.86-7.20) | 0.0317 |
| MR231 | PFD0520c | 6.03 (5.90-6.15) | 6.37 (6.22-6.52) | 0.0006 |
| MR232 | PFD0520c | 6.96 (6.82-7.11) | 7.50 (7.35-7.66) | 0 |
| MR232A | PFD0520c | 7.70 (7.57-7.83) | 8.26 (8.13-8.38) | 0 |
| MR233 | PFD0520c | 7.02 (6.90-7.13) | 7.46 (7.34-7.39) | 0 |
| AS182_15 | PFE0595w | 4.79 (4.64- 4.93) | 5.05 (4.89-5.20) | 0.0184 |
| MR246B | MAL6P1.37 | 5.79 (5.71-5.88) | 6.11(6.01-6.22) | 0 |
| MR259C | MAL6P1.37 | 6.88 (6.73-7.04) | 7.29 (7.13-7.46) | 0.0005 |
| MR260 | MAL6P1.37 | 6.24 (6.12-6.35) | 6.35( 6.18-6.51) | 0.2753 |
| MR282 (P27A) | MAL6P1.37 | 7.39 (7.26-7.52) | 7.86 (7.72-7.99) | 0 |
| MR261A | MAL6P1.37 | 6.71 (6.55- 6.87) | 7.22 (7.06-7.38) | 0 |
| LR170A | MAL8P1.12 | 7.16 (7.03-7.30) | 7.85 (7.71-8.00) | 0 |
| MR258 | MAL13P1.304 | 6.15 (5.98-6.32) | 6.80 (6.64-6.95) | 0 |
| MR214 | PFL0250w | 5.79 (5.64-5.93) | 6.21 (6.06-6.37) | 0.0001 |
| MR217 | PFL165w | 5.58 (5.47-5.69) | 6.13 (6.00-6.26) | 0 |
| MR217A | PFL165w | 6.20 (6.12-6.29) | 6.63 (6.54-6.72) | 0 |
| AS182.29 | PFL2310w | 6.13 (6.03-6.22) | 6.59 (6.48-6.69) | 0 |
| MR236 | PF08_0048 | 6.50 (6.35-6.65) | 6.98 (6.84-7.11) | 0 |
| MR236A | PF08_0048 | 6.30 (6.15-6.46) | 6.67 (6.53-6.82) | 0.001 |
| MR216 | PF11_0240 | 7.48 (7.42-7.54) | 7.74 (7.67- 7.81) | 0 |
| MR256 | PF13_0107 | 6.83 (6.73-6.93) | 7.43 (7.32-7.54) | 0 |
| LR162 | 6.57 (6.41- 6.73) | 6.85 (6.69-7.02) | 0.016 | |
| LR162A | 7.11 (6.96-7.26) | 7.43 (7.27- 7.59) | 0.0048 | |
| LR163 | PF11_0207 | 5.54 (5.34-5.74) | 6.94 (6.71-7.17) | 0 |
| MR198 | PF14_0089 | 7.78 (7.65-7.91) | 8.31 (8.17-8.44) | 0 |
| AS182_20 | PF14_0397 | 5.60 (5.49-5.72) | 5.71 (5.57- 5.85) | 0.2273 |
| MSP3 | 7.85 (7.67-8.03) | 8.18 (7.99-8.36) | 0.0153 | |
| MSP2_3D7 | 8.86 (8.73-8.99) | 8.81 (8.67-8.95) | 0.6125 | |
| (NANP)10 | 6.67 (6.54-6.81) | 8.27 (8.17-8.37) | 0 | |
Table 2: Comparisons of IgG level to the most reactive peptides between age groups.
Influence of the ethnicity on antigens reactivity and antibody intensity
We found that the total number of recognized peptides was higher in Fulani compared to mossi and Rimaibé ethnic groups. The same finding was observed about the total amount of IgG to reactive peptides.
The results related to the influence of the ethnicity on antibody reactivity profile and intensity are shown in Figure 2. When categorizing our study population in Fulani and non-Fulani, we observed that the Fulani recognized a higher number of peptides (65) compared with non-Fulani (59) the difference was statistically significant (<0.0001). The IgG level was also higher in Fulani than in the non-Fulani group (<0.0001).
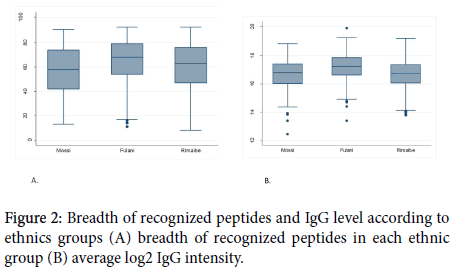
Figure 2. Breadth of recognized peptides and IgG level according to ethnics groups (A) breadth of recognized peptides in each ethnic group (B) average log2 IgG intensity.
The specific IgG response to each of the 36 most reactive peptides was also assessed. Of the 36 peptides, the IgG antibody response to 32 peptides was statically different between Fulani and the other sympatric ethnic groups with a higher response in Fulani. The results of this comparison are shown in Table 3.
| Name | Protein ID | Mean Intensity log IgG (95% CI) | ||
|---|---|---|---|---|
| Fulani | Non-Fulani | p-value | ||
| LR166 | PFA0635c | 5.98 (5.86-6.10) | 6.06 (5.96-6.15) | 0.3597 |
| LR146 | PFB0145c | 6.36 (6.14-6.57) | 6.59 (6.45-6.74) | 0.072 |
| LR149 | PFB0145c | 6.43 (6.26-6.59) | 6.03 (5.91-6.14) | 0.0001 |
| LR150A | PFB0145c | 7.20 (7.03-7.37) | 6.85 (6.69-7.00) | 0.0037 |
| LR150B | PFB0145c | 7.05 (6.85-7.24) | 6.75 (6.60-6.90) | 0.0191 |
| MR219 | PFB0315w | 6.79 (6.56-7.02) | 5.99 (5.81-6.18) | 0 |
| MR220 | PFC0345w | 7.29 (7.12-7.46) | 6.66 (6.53-6.79) | 0 |
| LR157A | PFD0110w | 7.14 (6.96-7.32) | 6.73 (6.58-6.88) | 0.0007 |
| MR231 | PFD0520c | 6.44 (6.29-6.60) | 6.00 (5.88-6.11) | 0 |
| MR232 | PFD0520c | 7.51 (7.35-7.67) | 6.99 (6.85-7.13) | 0 |
| MR232A | PFD0520c | 8.19 (8.03-8.35) | 7.77 (7.65-7.88) | 0 |
| MR233 | PFD0520c | 7.34 (7.21-7.47) | 7.11 (7.00-7.22) | 0.0131 |
| AS182_15 | PFE0595w | 5.01 (4.85-5.17) | 4.83 (4.69-4.97) | 0.1051 |
| MR246B | MAL6P1.37 | 6.04 (5.93-6.16) | 5.85 (5.76-5.93) | 0.0063 |
| MR259C | MAL6P1.37 | 7.46 (7.28-7.64) | 6.79 (6.65-6.94) | 0 |
| MR260 | MAL6P1.37 | 6.74 (6.58-6.89) | 6.00 (5.88-6.12) | 0 |
| MR282 (P27A) | MAL6P1.37 | 7.96 (7.81-8.10) | 7.35 (7.22-7.47) | 0 |
| MR261A | MAL6P1.37 | 7.38 (7.21-7.56) | 6.63 (6.48-6.78) | 0 |
| LR170A | MAL8P1.12 | 7.80 (7.66-7.95) | 7.23 (7.10-7.36) | 0 |
| MR258 | MAL13P1.304 | 6.65 (6.48-6.82) | 6.29 (6.13-6.45) | 0.0031 |
| MR214 | PFL0250w | 6.06 (5.90-6.22) | 5.90 (5.76-6.04) | 0.1532 |
| MR217 | PFL165w | 5.95 (5.79-6.10) | 5.71 (5.61-5.81) | 0.0091 |
| MR217A | PFL165w | 6.59 (6.49-6.70) | 6.24 (6.16-6.32) | 0 |
| AS182.29 | PFL2310w | 6.40 (6.29-6.52) | 6.25 (6.16-6.35) | 0.0493 |
| MR236 | PF08_0048 | 6.88 (6.73-7.04) | 6.59 (6.45- 6.73) | 0.0063 |
| MR236A | PF08_0048 | 6.58 (6.41-6.75) | 6.39 (6.24-6.53) | 0.0883 |
| MR216 | PF11_0240 | 7.73 (6.66-7.79) | 7.50 (7.44-7.56) | 0 |
| MR256 | PF13_0107 | 7.29 (7.17-7.4) | 6.93 (6.84-7.03) | 0 |
| LR162 | 6.99 (6.82-7.17) | 6.49 (6.33-6.64) | 0 | |
| LR162A | 7.49 (7.32-7.66) | 7.09 (6.95-7.23) | 0.0006 | |
| LR163 | PF11_0207 | 7.16 (6.94-7.38) | 5.44 (5.24-5.64) | 0 |
| MR198 | PF14_0089 | 8.30 (8.16-8.43) | 7.81 (7.68-7.93) | 0 |
| AS182_20 | PF14_0397 | 5.90 (5.74-6.05) | 5.49 (5.38-5.59) | 0 |
| MSP3 | 8.14 (7.94-8.34) | 7.90 (7.73-8.07) | 0.0713 | |
| MSP2_3D7 | 9.25 (9.13-9.38) | 8.59 (8.46-8.72) | 0 | |
| (NANP)10 | 7.81 (7.66-7.96) | 7.03 (6.90-7.16) | 0 | |
Table 3: Comparisons of IgG level to the most reactive peptides between Ethnic groups.
Influence of the hemoglobin type and gender on antigens reactivity and antibody intensity
The average number of recognized peptides was similar (62) in volunteers with hemoglobin AA and non-AA (p=0,6439). The same finding was observed concerning the total amount of IgG between the two groups (p=0,3448).
Concerning the specific IgG response to the most reactive peptides; there was not statistically a significant difference between hemoglobin AA carriers and non-AA to 32 of the most reactive peptides. The results related to the influence of hemoglobin type and sex on peptides reactivity and the IgG level are shown in Figures 3 and 4.
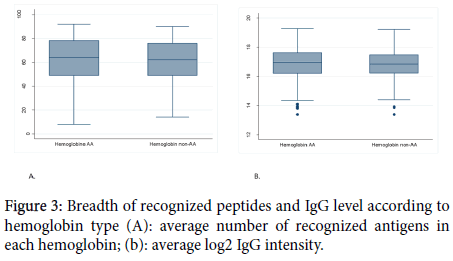
Figure 3. Breadth of recognized peptides and IgG level according to hemoglobin type (A): average number of recognized antigens in each hemoglobin; (b): average log2 IgG intensity.
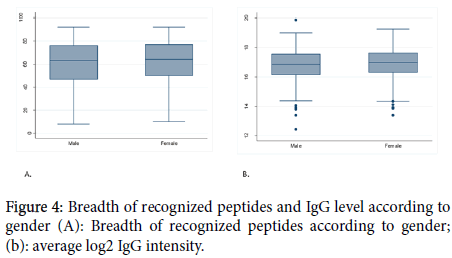
Figure 4. Breadth of recognized peptides and IgG level according to gender (A): Breadth of recognized peptides according to gender; (b): average log2 IgG intensity.
The influence of hemoglobin on the specific IgG antibody response to each of the 36 most reactive antigens is reported in Table 4 while Table 5 reports the comparison of Ig response between male and female.
| Name | Protein ID | Mean Intensity log IgG (95% CI) | ||
|---|---|---|---|---|
| Hemoglobin AA | Hemoglobin non-AA | p-value | ||
| LR166 | PFA0635c | 6.062 (5.97- 6.15) | 5.95 (5.80-6.10) | 0.243 |
| LR146 | PFB0145c | 6.47 (6.32-6.61) | 6.65 (6.41-6.88) | 0.2195 |
| LR149 | PFB0145c | 6.20 (6.08-6.32) | 6.11 (5.92-6.31) | 0.4514 |
| LR150A | PFB0145c | 7.06 (6.93- 7.19) | 6.86 (6.59-7.14) | 0.1587 |
| LR150B | PFB0145c | 6.87 (6.72-7.02) | 6.88 (6.64-7.12) | 0.9281 |
| MR219 | PFB0315w | 6.34 (6.16- 6.51) | 6.21 (5.91-6.52) | 0.493 |
| MR220 | PFC0345w | 6.913 (6.78-7.03) | 6.83 (6.60-7.06) | 0.5696 |
| LR157A | PFD0110w | 6.94 (6.80- 7.09) | 6.81 (6.56-7.06) | 0.3416 |
| MR231 | PFD0520c | 6.19 (6.07-6.30) | 6.06 (5.86-6.26) | 0.2666 |
| MR232 | PFD0520c | 7.32 (7.19- 7.44) | 6.85 (6.61- 7.09) | 0.0004 |
| MR232A | PFD0520c | 7.97 (7.85-8.08) | 7.77 (7.58-7.96) | 0.0884 |
| MR233 | PFD0520c | 7.24 (7.14-7.34) | 7.07 (6.88-7.26) | 0.1024 |
| AS182_15 | PFE0595w | 4.93 (4.80- 5.06) | 4.73 (4.53-4.94) | 0.7586 |
| MR246B | MAL6P1.37 | 5.91 (5.83-5.99) | 5.89 (5.74-6.04) | 0.8134 |
| MR259C | MAL6P1.37 | 7.09 (6.96-7.23) | 6.99 (6.73-7.24) | 0.4378 |
| MR260 | MAL6P1.37 | 6.29 (6.18-6.41) | 6.22 (6.01-6.42) | 0.5086 |
| MR282 (P27A) | MAL6P1.37 | 7.60 (7.48-7.72) | 7.54 (7.35-7.73) | 0.6084 |
| MR261A | MAL6P1.37 | 6.92 (6.78-7.06) | 6.94 (6.71-7.17) | 0 |
| LR170A | MAL8P1.12 | 7.46 (7.34-7.59) | 7.38 (7.18-7.57) | 0.4675 |
| MR258 | MAL13P1.304 | 6.49 (6.35-6.63) | 6.35 (6.08-6.62) | 0.3502 |
| MR214 | PFL0250w | 6.00 (5.87-6.13) | 5.94 (5.72-6.16) | 0.6582 |
| MR217 | PFL165w | 5.81 (5.70-5.91) | 5.80 (5.62-5.97) | 0.9167 |
| MR217A | PFL165w | 6.40 (6.32-6.47) | 6.26 (6.12-6.40) | 0.0862 |
| AS182.29 | PFL2310w | 6.30 (6.21-6.40) | 6.37 (6.24-6.50) | 0.4455 |
| MR236 | PF08_0048 | 6.75 (6.63-6.87) | 6.56 (6.34- 6.78) | 0.1382 |
| MR236A | PF08_0048 | 6.50 (6.37-6.62) | 6.26 (6.00-6.52) | 0.077 |
| MR216 | PF11_0240 | 7.58 (7.52-7.63) | 7.60 (7.50- 7.69) | 0.7586 |
| MR256 | PF13_0107 | 7.10 (7.00-7.19) | 7.04 (6.89-7.19) | 0.5236 |
| LR162 | 6.73 (6.59-6.87) | 6.67 (6.44-6.90) | 0.671 | |
| LR162A | 7.27 (7.14-7.40) | 7.23 (7.01- 7.46) | 0.7703 | |
| LR163 | PF11_0207 | 6.30 (6.10-6.49) | 5.91 (5.59-6.24) | 0.0513 |
| MR198 | PF14_0089 | 8.07 (7.96-8.18) | 7.86 (7.67-8.05) | 0.0665 |
| AS182_20 | PF14_0397 | 5.67 (5.56-5.78) | 5.57 (5.39- 5.76) | 0.3901 |
| MSP3 | 8.04 (7.87-8.18) | 7.90 (7.59-8.20) | 0.3973 | |
| MSP2_3D7 | 8.90 (8.79-9.02) | 8.64 (8.43-8.85) | 0.0272 | |
| (NANP)10 | 7.24 (7.11-7.37) | 7.54 (7.37-7.72) | 0.0138 | |
Table 4: Comparisons of IgG level to the most reactive peptides according to Hemoglobin type.
| Name | Protein ID | Mean Intensity log IgG (95% CI) | ||
|---|---|---|---|---|
| Male | Female | p-value | ||
| LR166 | PFA0635c | 6.041 (5.93-6.14) | 6.02 (5.91- 6.12) | 0.7735 |
| LR146 | PFB0145c | 6.46 (6.28-6.63) | 6.55 (6.39-6.72) | 0.437 |
| LR149 | PFB0145c | 6.12 (5.98-6.26) | 6.23 (6.10-6.36) | 0.2625 |
| LR150A | PFB0145c | 6.95 (6.79-7.12) | 7.02 (6.86-7.17) | 0.6067 |
| LR150B | PFB0145c | 6.83 (6.65-7.01) | 6.90 (6.74- 7.07) | 0.5495 |
| MR219 | PFB0315w | 6.38 (6.17-6.59) | 6.21 (6.01-6.41) | 0.2487 |
| MR220 | PFC0345w | 6.95 (6.80- 7.10) | 6.85 (6.70-7.00) | 0.3574 |
| LR157A | PFD0110w | 6.88 (6.71-7.05) | 6.89 (6.74-7.05) | 0.9163 |
| MR231 | PFD0520c | 6.14 (6.01-6.28) | 6.18 (6.04-6.31) | 0.7087 |
| MR232 | PFD0520c | 7.16 (7.00-7.32) | 7.22 (7.07-7.37) | 0.5702 |
| MR232A | PFD0520c | 7.90 (7.76-8.04) | 7.94 (7.82-8.07) | 0.6585 |
| MR233 | PFD0520c | 7.17 (7.04-7.29) | 7.22 (7.10-7.34) | 0.5408 |
| AS182_15 | PFE0595w | 4.87 (4.70-5.03) | 4.93 (4.79-5.07) | 0.5473 |
| MR246B | MAL6P1.37 | 5.95 (5.85-6.05) | 5.89 (5.80-5.98) | 0.3866 |
| MR259C | MAL6P1.37 | 6.94 (6.77-7.11) | 7.16 (7.01-7.31) | 0.0557 |
| MR260 | MAL6P1.37 | 6.28 (6.14-6.42) | 6.27 (6.14-6.41) | 0.9171 |
| MR282 (P27A) | MAL6P1.37 | 7.47 (7.33-7.61) | 7.67 (7.53-7.80) | 0.0487 |
| MR261A | MAL6P1.37 | 6.84 (6.66-7.02) | 7.00 (6.85- 7.15) | 0.1809 |
| LR170A | MAL8P1.12 | 7.40 (7.25-7.55) | 7.51 (7.38-7.64) | 0.2858 |
| MR258 | MAL13P1.304 | 6.51 (6.33-6.68) | 6.39 (6.22-6.55) | 0.3155 |
| MR214 | PFL0250w | 6.02 (5.85- 6.17) | 5.92 (5.77- 6.06) | 0.3709 |
| MR217 | PFL165w | 5.76 (5.63-5.89) | 5.84 (5.73-5.95) | 0.367 |
| MR217A | PFL165w | 6.35 (6.26-6.44) | 6.39 (6.30-6.47) | 0.5357 |
| AS182.29 | PFL2310w | 6.30 (6.19-6.40) | 6.32 (6.22-6.42) | 0.7494 |
| MR236 | PF08_0048 | 6.67 (6.51-6.82) | 6.74 (6.60-6.88) | 0.4903 |
| MR236A | PF08_0048 | 6.41 (6.24-6.58) | 6.51 (6.37-6.65) | 0.3603 |
| MR216 | PF11_0240 | 7.57 (7.50-7.63) | 7.60 (7.53-7.66) | 0.563 |
| MR256 | PF13_0107 | 7.00 (6.89-7.11) | 7.13 (7.02-7.23) | 0.1145 |
| LR162 | 6.59 (6.42-6.77) | 6.78 (6.63-6.94) | 0.1083 | |
| LR162A | 7.19 (7.02- 7.35) | 7.29 (7.15-7.44) | 0.3386 | |
| LR163 | PF11_0207 | 5.94 (5.71-6.18) | 6.37 (6.15- 6.58) | 0.0097 |
| MR198 | PF14_0089 | 8.01 (7.88-8.13) | 7.99 (7.85-8.12) | 0.828 |
| AS182_20 | PF14_0397 | 5.67 (5.54-5.79) | 5.63 (5.50-5.75) | 0.6726 |
| MSP3 | 7.96 (7.77-8.15) | 8.03 (7.85-8.21) | 0.5831 | |
| MSP2_3D7 | 8.77 (8.62-8.91) | 8.91 (8.78-9.04) | 0.1438 | |
| (NANP) 10 | 7.20 (7.06-7.34) | 7.44 (7.29-7.58) | 0.02 | |
Table 5: Comparisons of IgG level to the most reactive peptides according to sex.
Kinetics of the IgG antibody response acquisition to the top 5 most reactive alpha-helical coiled coil segments
The concomitant influence of age and ethnicity was also investigated to explore the trend of the acquisition of the antibody response over time and between ethnic groups. For this purpose, we have selected the 5 most reactive alpha-helical coiled coil segments and MSP2 as control as shown in Figure 5.
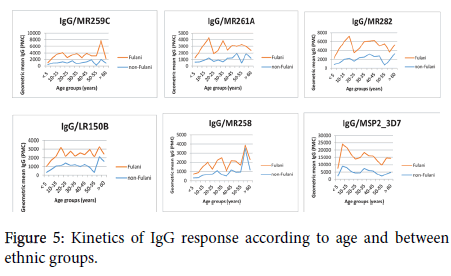
Figure 5. Kinetics of IgG response according to age and between ethnic groups.
Our findings showed that the acquisition of the antibody increase over time for both Fulani and non-Fulani individuals. In the Fulani group, the maximum of IgG response was observed in people whose age was between 15 and 20 years old for some antigens such as MR261A, MR282, LR150B, and MSP2. For the same antigens, the maximum of IgG in non-Fulani individuals was observed in people older than 40 years old.
Discussion
In this study, we investigated the influence of age, ethnicity, hemoglobin type and sex on the acquisition of immunity to Plasmodium falciparum alpha-helical coiled-coil proteins segments in populations living in a malaria-endemic area. The knowledge of the influence of those factors on the acquisition of immunity against malaria is essential in the selection of Plasmodium antigens as potential malaria vaccine candidates. We have used protein microarrays technology, a high throughput immunoassay that gives the possibility to screen in parallel many antigens in a single experiment. Some previous studies conducted in the same communities, which were involved in our studies, showed the influence of some factors such as age and ethnicity on the acquisition of the immunity against some malaria antigens. In our study, in addition to age and ethnicity, we have investigated whether the hemoglobin type and also the gender are factors that influence the acquisition of immunity in our study population.
The data obtained from our study demonstrate that the breadth and the magnitude of the IgG antibody responses to Plasmodium falciparum alpha-helical coiled coil antigens were higher in adults compared to adolescents and children. These findings confirm the results obtained with other malaria antigens that age has an influence in the acquisition of immunity in populations living in a malariaendemic area. Previous studies showed that there is a positive correlation between age and the acquisition of immunity to malaria in people living in endemic areas [6-8].
With regard to the influence of the ethnicity on the acquisition of the immunity, our study shows that among the 3 ethnic populations, the Fulani group appears to have the highest number of recognized antigens and also the highest intensity of IgG antibodies compared to Mossi and Rimaibe confirming previous findings [15,16]. Additional studies conducted in Mali also showed that the Fulani was more responders than other sympatric ethnic populations [17,18].
Genetic studies based on some classical genes that confer resistance to malaria were explored to understand the high response of Fulani compared to the sympatric ethnic groups. The findings of that study showed that the Fulani groups had a low prevalence of these genes compared to the sympatric groups [19,20]. More recently, a study conducted in the same communities showed that transcriptional changes were observed in Fulani and could explain the lower susceptibility to malaria infection of this group compared to that of other populations [20]. We have also investigated the influence of hemoglobin in the acquisition of the immunity to the antigens used in our study. Several studies showed evidence related to the role of hemoglobin S and C in the protection against severe and uncomplicated malaria [21,22]. The role of hemoglobin S and C has already shown in the protection against malaria infection [12].
To investigate the influence of hemoglobin S and C on the acquisition of acquired immunity, the study populations were categorized in 2 groups: the group of volunteers whose hemoglobin genotype was AA and the group of volunteers with a least one allele of abnormal hemoglobin (AS, AC, CC, and SC) considered as non-AA. We found that in general hemoglobin type did not influence the breadth and the magnitude of IgG antibody in our study population except for MSP2_3D7 and (NANP)10. A study conducted in Mali showed that the hemoglobin type did not influence the antibody responses [23]. One hypothesis is that HbAS and HbAC accelerate the acquisition of immunity to malaria, possibly by enhancing Plasmodium falciparum-specific antibody responses. However, in that study, protein microarray data showed that there were no significant differences in antibody profiles between normal and sickle-cell trait. Another study conducted in Burkina Faso has shown that the naturally acquired antibody response to some malaria antigens was higher in hemoglobin S and C carriers [24]. We also investigated whether the acquisition of immunity is influenced by gender. For this purpose, we have compared the average number of recognized antigens and also the intensity of the IgG antibody in male and female groups. We found that neither the breadth nor the intensity of the IgG antibody response was influenced by gender. We selected the peptides with the most positive reactivity prevalence and good intensity and determine the influence of the factors mentioned above in the acquisition of antigenspecific IgG antibody response. In general, for most of them, the influence of age and ethnicity are confirmed but not for hemoglobin type and sex.
Taking into account the same antigens, people from the Fulani group seem to reach earlier the maximum of IgG response to antigens compared with the individuals of the non-Fulani group.
Conclusion
The findings from this study confirm that a high throughput protein microarrays immunoassay is applicable to Plasmodium falciparum alpha-helical coiled-coil proteins segments to explore the naturally acquired immunity for the discovery of new malaria antigens. Age and ethnicity are found to influence the IgG antibody reactivity and intensity to the proteins tested. Other factors such as hemoglobin type and sex did not have a significant influence on the acquired IgG antibody reactivity and intensity. These findings should be considered in any strategy for new antigen related to alpha-helical coiled coil protein segments discovery for a potential vaccine clinical development.
Acknowledgment
We are grateful to all the study participants who have made this study possible and also to all the staff of CNRFP who have contributed to the work.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was financially supported by the Italian cooperation in Burkina Faso.
Author Contributions
OO, YT, GC, VM, DM, SBS, RS, IN: Conceived and design the experiment; OO, LN, ECB, AD, YK: Performed the experiments; OO, BK, ABT, IN: Analyzed the data; OO, LN, ECB, YK, DA, BT, ABT, GC, VM, DM, YT, SBS, RS, IN: contributed in writting of the manuscript.
REFERENCES
- WHO (2018) World malaria report 2018.
- Gustavo CC, Ian HC, Steve FS, Shalini N, Marina McDW, Aung PP, et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017;18:78.
- Jacob MR, Sulaiman SI, Charles M, Rousseau D, Helen I, Murielle JW, et al. Genome-wide transcription and functional analyses reveal heterogeneous molecular mechanisms driving pyrethroids resistance in the major malaria vector anopheles funestus across Africa. G3-GENES GENOM GENET. 2017;7:1819-1832.
- Baird JK, Masbar S, Basri H, Tirtokusumo S, Subianto B, Hoffman SL, et al. Age-dependent susceptibility to severe disease with primary exposure to Plasmodium falciparum. J Infect Dis. 1998;178:592-595.
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498-511.
- Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE , et al. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci. 2010;107:6958-6963.
- Baum E, Badu K, Molina DM, Liang X, Felgner PL, Yan G, et al. Protein microarray analysis of antibody responses to Plasmodium falciparum in western Kenyan highland sites with differing transmission levels. PLoS One. 2013;8:e82246.
- Dent AE, Nakajima R, Liang L, Baum E, Moormann AM, Sumba PO, et al. Plasmodium falciparum protein microarray antibody profiles correlate with protection from symptomatic malaria in Kenya. J Infect Dis. 2015;212:1429-1438.
- Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680-4694.
- Gray JC, Corran PH, Mangia E, Gaunt MW, Li Q, Tetteh KK, et al. Profiling the antibody immune response against blood stage malaria vaccine candidates. Clin Chem. 2007;53:1244-1253.
- Villard V, Agak GW, Frank G, Jafarshad A, Servis C, Nébié I, et al. Rapid identification of malaria vaccine candidates based on alpha-helical coiled coil protein motif. PLoS One. 2007;2:e645.
- Mangano VD, Kabore Y, Bougouma EC, Verra F, Sepulveda N, Bisseye C, et al. Novel insights into the protective role of hemoglobin s and c against Plasmodium falciparum parasitemia. J Infect Dis. 2015;212:626-634.
- Ross P, Hall L, Smirnov I, Haff L. High-level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:1347-1351.
- Olugbile S, Kulangara C, Bang G, Bertholet S, Suzarte E, Villard V, et al. Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein PFF0165c. Infect Immun. 2009;77:5701-5709.
- Modiano D, Petrarca V, Sirima BS, Nebie I, Diallo D, Esposito F, et al. Different response to Plasmodium falciparum malaria in west African sympatric ethnic groups. Proc Natl Acad Sci. 1996;93:13206-13211.
- Modiano D, Chiucchiuini A, Petrarca V, Sirima BS, Luoni G, Perlmann H, et al. Humoral response to Plasmodium falciparum Pf155/ring-infected erythrocyte surface antigen and Pf332 in three sympatric ethnic groups of Burkina Faso. Am J Trop Med Hyg. 1998;58:220-224.
- Dolo A, Modiano D, Maiga B, Daou M, Dolo G, Guindo H, et al. Difference in susceptibility to malaria between two sympatric ethnic groups in Mali. Am J Trop Med Hyg. 2005;72:243-248.
- Arama C, Maiga B, Dolo A, Kouriba B, Traore B, Crompton PD, et al. Ethnic differences in susceptibility to malaria: what have we learned from immuno-epidemiological studies in West Africa? Acta Trop. 2015;146:152-156.
- Modiano D, Luoni G, Sirima BS, Lanfrancotti A, Petrarca V, Cruciani F, et al. The lower susceptibility to Plasmodium falciparum malaria of Fulani of Burkina Faso (West Africa) is associated with low frequencies of classic malaria-resistance genes. Trans Royal Soc Trop Med Hyg. 2001;95:149-152.
- Quin JE, Bujila I, Cherif M, Sanou GS, Qu Y, Vafa HM, et al. Major transcriptional changes observed in the Fulani, an ethnic group less susceptible to malaria. Elife 6. 2017.
- Modiano D, Luoni G, Sirima BS, Simpore J, Verra F, Konaté A, et al. Haemoglobin C protects against clinical Plasmodium falciparum malaria. Nature. 2001;414:305-308.
- Mockenhaupt FP, Ehrhardt S, Cramer JP, Otchwemah RN, Anemana SD, Goltz K, et al. Hemoglobin C and resistance to severe malaria in Ghanaian children. J Infect Dis. 2004;190:1006-1009.
- Tan X, Traore B, Kayentao K, Ongoiba A, Doumbo S, Waisberg M, et al. Hemoglobin S and C heterozygosity enhances neither the magnitude nor breadth of antibody responses to a diverse array of Plasmodium falciparum antigens. J Infect Dis. 2011;204:1750-1761.
- Verra F, Simpore J, Warimwe GM, Tetteh KK, Howard T, Howard T, et al. Haemoglobin C and S role in acquired immunity against Plasmodium falciparum malaria. PLoS One. 2007;2:e978.
Citation: Ouedraogo O, Nunziangeli L, Bougouma EC, Kabor Y, Diarra A, et al. (2019) Seroreactivity of Populations Living in Endemic Area of Burkina Faso to Plasmodium falciparum Alpha-Helical Coiled Coil Proteins Motifs by Protein Microarray. J Trop Dis 7: 320. doi: 10.35248/2329-891X.19.7.320
Copyright: © 2019 Ouedraogo O, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.