Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2020) Volume 11, Issue 8
Screening of Chickpea (Cicer arietinum) Genotypes to Three Identified Races of Fusarium oxysporum f. sp. ciceris in Sudan
Omyma Elmahi Mohamed1* and Amel Adam Mohamed22Chickpea Breeding Program, Agricultural Research Corporation, Sudan
Received: 07-Jun-2020 Published: 12-Aug-2020, DOI: 10.35248/2157-7471.20.11.507
Abstract
Fusarium wilt is the main biotic stress that limited chickpea production in Sudan. Twenty chickpea genotypes were screened for resistance against Fusarium oxysporum f. sp. ciceris (Foc) the causal agent of Fusarium wilt of chickpea. The study was done in pot experiment at Gezira Research Station greenhouse. The genotypes were screened against three identified races of Foc in Sudan namely race 0, 2 and unidentified race to know their reaction against the pathogen.Race 0 is the most prevalent and widely spread in Sudan, while the unidentified race is limited to Gezira State. The variety Hawta (Iccv-92318) showed resistant reaction to the three tested Foc races while Shendi (ILC-1335) and Jabel Marra (ILC-915) varieties showed susceptible reaction to race 0 and highly susceptible reaction to the other two races. The other genotypes showed susceptible reactions to the unidentified race and variable reactions to races 2 and 0. This evaluation has helped to identify new sources of resistance to wilt disease for use in chickpea breeding program and for proper chickpea cultivation in the different areas of Sudan. It also will be useful in preventing the spread of the pathogen from infested areas to uninfected ones.
Keywords
Cicer arietinum; Fusarium oxysporum; Resistance; Screening; Sudan
Introduction
Chickpea (Cicer arietinum L) is a cool season food legume and an important cash crop in Sudan contributing in the sustainability and profitability of production systems. The crop was traditionally grown on residual moisture of the flood in northern Sudan [1] and recently it is introduced to the central parts of the country as an irrigated crop. Fusarium wilt disease caused by the fungus Fusarium oxysporum Schlechtend. Fusarium oxysporum f. sp. ciceris (Padwick) Matuo & K. Sato (Foc) is one of the most important vascular wilt diseases of the crop. The disease emerged as a devastating and economically important constraint that affects chickpea production in Sudan. The fungus could survive on crop residues in soil for more than 6 years thus the practical and cost-efficient method for management is the use of resistant cultivars which represent a key component in integrated disease management (IDM) programs [2].
A field survey was conducted during seasons 2011-2012 in the main chickpea production areas of Sudan to determine the importance and distribution of wilt and root rot disease [3] and to investigate the genetic diversity of Sudanese Foc isolates [4]. These studies revealed that management of chickpea wilt and root rot disease should be done through integrated crop management approach by using high yielding, wilt/root rot resistant cultivars with recommended seeding rates, irrigation frequency, sowing dates and appropriate cropping sequences [3].
The Sudanese isolates collected from six different chickpeagrowing states in the Sudan were characterized using four random amplified polymorphic DNA (RAPD), three simple sequence repeats (SSR), five sequence-characterized amplified region (SCAR) primers and three specific Foc genome primers. Based on the similarity coefficient, the results indicated that the isolates from the Sudan were grouped and identified as 0, 2 and unknown races. This study identified a new race of Foc (race 0) in the Sudan [4]. Race 0 is widely distributed in central Sudan, while the unidentified race is limited to Gezira State. This clearly indicates that race 0 is the most prevalent and widely spread. Race 2 is prevalent in Northern State, River Nile State and northern parts of Gezira State.
Adequate characterization of the resistance of chickpea lines and cultivars to specific races of F. oxysporum f. sp. ciceris is essential for resistance deployment. In the absence of agronomical and/or commercially suitable resistant cultivars, prediction of disease risk potential in a geographic area based on assessment of pathogen race and inoculum density thresholds in soil and susceptibility of cultivars can be of use for the management of Fusarium wilt in chickpea [5]. Thus the current study was carried out to assess the reaction of the released chickpea cultivars and some other promising genotypes in Sudan to the recently identified races of Foc.
Materials and Methods
Twenty chickpea genotypes (8 released varieties + 12 genotypes) (Table 1) were screened to determine their reaction to the identified races 0, 2 and X (unidentified) [4] under greenhouse conditions. Inoculums from pure single spore cultures of each of the three races were multiplied on PDA media. With a 3-mm cork borer, 2 agar plugs were used to inoculate and increased in 180 g Sorghum Sand Mixture (SSM =135 g sorghum + 45 g sand + 100 ml water) autoclaved at 121°C for 20 minutes in 500-ml flasks (21- 1). The inoculated flasks were incubated at 25°C and a 12-h photoperiod using fluorescent lamps for two weeks. The infested sorghum sand mixture was mixed thoroughly with 4 kg sterilized soil (clay loam/sand, 1: 1, w/w). Surface sterilized ten seeds of each genotype were sown in a separate pot filled with infested SSM. The pots were arranged in a completely randomized design with 2 replicates for each race-genotype combination and were kept in the greenhouse under natural conditions. The plants were observed daily for symptoms development. Disease reactions were assessed and scored four and six weeks after inoculation. The level of resistance and susceptibility of each tested genotype was determined by using the rating scale of Iqbal et al., [6] and Suliman [7] with some modifications where R=0-20% wilted plants=1, MR=21-40% wilted plants=2, S=41-80% wilted plants=3 and HS = ≥ 80% wilted plants=4. Wilted plants were checked for vascular discoloration symptoms and re-isolation to confirm that the disease is caused by Foc.
| S. No | Accession No. | Name | No | Accession No. |
|---|---|---|---|---|
| Released cultivars | 9 | Elixir | ||
| 1 | ILC 915 | Jabel Marra | 10 | FLIP 93-93 |
| 2 | ILC 1335 | Shendi | 11 | ILC 1929 |
| 3 | Iccv-92318 | Hwata | 12 | FLIP 97-263c |
| 4 | ICCV-2 | Wad Hamid | 13 | ILC 464 |
| 5 | Iccv-91302 | Burgeig | 14 | FLIP 84-79c |
| 6 | FLIPc 82-89 | Salwa | 15 | FLIP 81-71c |
| 7 | FLIP 77-91c | Matama | 16 | FLIP 97-530 |
| 8 | Iccv-89509 | Atmor | 17 | FLIP 03-104c |
| 18 | ILC 3279 | |||
| 19 | FLIP 84-48c | |||
| 20 | FLIP 85-17c | |||
Table 1: List of chickpea screened genotypes.
The data was analyzed using MSTSC Program and the Duncan Multiple Range Test was used for mean separation.
Results
There were significant differences between genotypes for the different studied races (Table 2). Figure 1 illustrated the variable virulence of the three races on the tested chickpea genotypes. Race 0 isolates showed progressive yellowing of the plants and wilting within 45 days after inoculation, whereas race 2 isolates exhibited wilting of the plants after one month from inoculation. Race X (unidentified) showed wilting of the plants within 45 days after inoculation without yellowing. Symptoms of yellowing and wilting plants as compared with healthy plants were exhibited in Figure 2. Jabel Marra, Shendi, Elixir and FLIP 84-48c were the only genotypes susceptible to race 0. All other genotypes were either resistant or moderately resistant to the same race. All genotypes except of Hwata variety were susceptible to the unidentified race (X). However, Jabel Marra and Shendi varieties were highly susceptible to this race. The genotypes Jabel Marra, Shendi, Wad Hamid, Salwa, Matama, Atmoor, ILC 464, FLIP 84-79C, FLIP-03-104c and FLIP 84-48c were susceptible or highly susceptible to isolates of race 2. Hwata variety reaction to the three tested races was either resistant or moderately resistant (Table 2 and Figures 1 and 2).
| No. | Genotypes | Grade | Reaction to R 0 | Grade | Reaction to R X | Grade | Reaction to R 2 |
|---|---|---|---|---|---|---|---|
| 1 | JabelMarra | 3.33 b | S | 4.00 AM | HS | 3.67 a | HS |
| 2 | Shendi | 2.67 c | S | 4.00 AM | HS | 3.33 ab | S |
| 3 | Hwata | 1.67 d | MR | 1.00 c | R | 2.00 cd | MR |
| 4 | Wad Hamid | 1.33 de | R | 3.00 b | S | 3.67 a | HS |
| 5 | Burgeig | 1.33 de | R | 3.00 b | S | 1.33 d | R |
| 6 | Salwa | 1.33 de | R | 3.00 b | S | 3.67 a | HS |
| 7 | Matama | 1.33 de | R | 3.00 b | S | 3.67 a | HS |
| 8 | Atmor | 1.67 c | MR | 4.00 AM | HS | 3.67 a | HS |
| 9 | Elixir | 3.00 bc | S | 3.33 b | S | 1.33 d | R |
| 10 | FLIP 93-93 | 1.00 e | R | 3.33 b | S | 1.67 d | MR |
| 11 | ILC 1929 | 1.33 de | R | 3.33 b | S | 1.67 d | MR |
| 12 | FLIP 97-263c | 1.67 d | MR | 3.33 b | S | 1.67 d | MR |
| 13 | ILC 464 | 1.67 d | MR | 3.33 b | S | 2.67 bc | S |
| 14 | FLIP 84-79c | 1.67 d | MR | 3.33 b | S | 2.67 bc | S |
| 15 | FLIP 81-71c | 1.67 d | MR | 3.33 b | S | 2.00 cd | MR |
| 16 | FLIP 97-530 | 1.00 e | R | 3.33 b | S | 1.67 d | MR |
| 17 | FLIP 03-104c | 1.00 e | R | 3.33 b | S | 3.00 ab | S |
| 18 | ILC 3279 | 1.00 e | R | 3.33 b | S | 1.67 d | MR |
| 19 | FLIP 84-48c | 4.00 AM | HS | 3.33 b | S | 3.00 ab | S |
| 20 | FLIP 85-17c | 1.33 de | R | 3.33 b | S | 1.67 d | MR |
| CV% | 18.98 | 8.93 | 22.09 | ||||
| SE± | 0.197 | 0.168 | 0.317 |
Table 2: Reaction of chickpea genotypes to Race 0, Race 2 and the unidentified race of Fusarium oxysporum f. sp. ciceris.
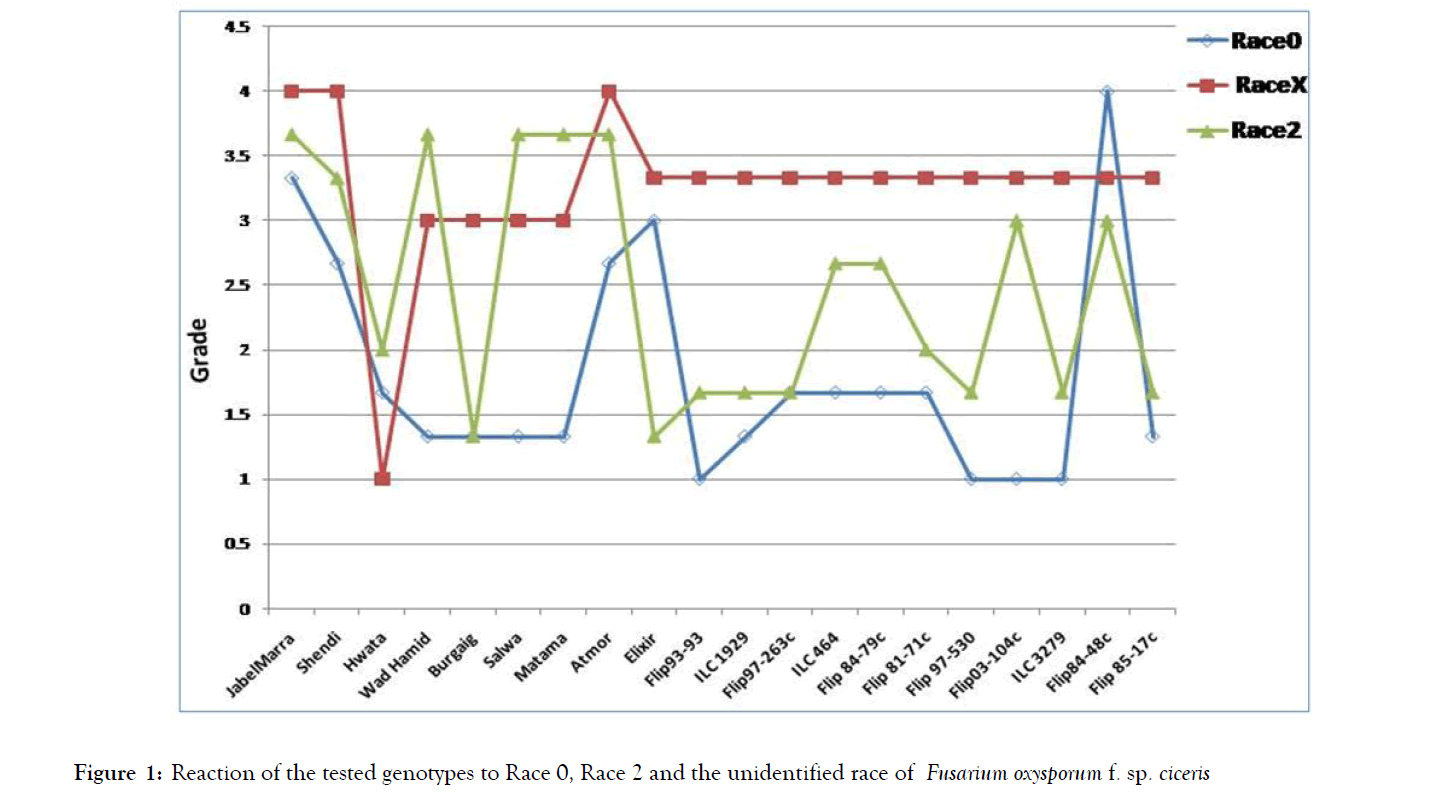
Figure 1: Reaction of the tested genotypes to Race 0, Race 2 and the unidentified race of Fusarium oxysporum f. sp. ciceris.
Figure 2:Symptoms of yellowing and wilting of plants as compared with healthy plants. (1= Healthy plants, 2= yellowing of the lower leaves, 3= Wilted plants).
Discussion
Chickpea production was affected by wilt diseases in all States of Sudan with varying incidence levels which was affected by chickpea cultivars, soil types, and agronomic and cropping system practices. Large number of germplasm lines were introduced to Sudan and by extensive screening and selection methods between breeders and pathologists at River Nile State, a number of lines emerged and tested [8]. Selections were for adaptation, high and stable yield, resistance to wilt/root-rot disease, earliness and large seed size. The evaluation justified the release of ten cultivars with varying degrees of resistance to wilt and root rot disease. Actually, the effectiveness of resistant cultivars is limited by the occurrence of different pathotypes and races of the fungus as the crop was entered to the central States of the country.
The screening of the chickpea genotypes against the identified races (0, 2 and X), demonstrated that the already released cultivars (Shendi and Jabel Marra) were susceptible to the three identified races of the pathogen. If we need to save Shendi and Jabel Marra as high yielding and good quality chickpea cultivars, incorporation of the resistant genes of Fusarium wilt pathogen to those cultivars becomes indispensable. Hawta variety is a promising variety resistant to the three races and could be introduced in demonstration fields to chickpea farmers. Hwata and Burgeig varieties were identified as the top seed yielding varieties that adapted to favorable conditions of Sudan [9]. Furthermore, Jabel Marra variety gave high seed yield and recognized as optional under poor soil at high terrace [10]. The released varieties Wad Hamid, Burgeig, Salwa, Matama and Atmor have fluctuating reactions to the three races. All the tested cultivars and lines are susceptible to the unidentified race found in Sudan and this indicates that it is the most aggressive race in Sudan (Figure 1). To overcome this situation in areas where this race is prevailing, a large chickpea germplasm should be screened for resistance to this race. Similar results of variation in chickpea resistance were reported in Sudan, India, Pakistan, Italy, Syria and Tunisia [11-18]. Also the greenhouse screening of chickpea should be extended to field testing to confirm resistance in the field. Therefore, the stage of plant growth at which infection occurs is crucial as the symptoms of the disease started very late after 45 days from sowing in case of race 0 and the unidentified one. Race 0 is widely distributed in central states of Sudan, while the unidentified race is limited to Gezira State. This clearly indicates that race 0 is the most prevalent and widely spread. Therefore, some measures should be taken to prevent its spread to other new areas.
Sources of resistance against F. oxysporum f. sp. ciceris have been identified [19,20] and exploited in several chickpea breeding programs consequently. Fair number of resistant chickpea germplasm lines operative against specific races of the pathogen has been developed globally.
Conclusion
In conclusion, there should be a breeding program to replace the highly susceptible varieties by resistant one for the identified races of Foc. The resistant Hwata variety should be shown in the disease infected areas to provide high and stable yield. Certified seeds of the promising resistant genotypes should be produced and distributed to farmers for commercial production to replace the wilt susceptible chickpea varieties. There is a need to conduct surveys to understand the presence of different pathotypes of Foc since knowledge on variability of pathogen is essential in breeding for durable resistance and would overcome the challenge of breakdown of resistance due to development of new pathotypes.
REFERENCES
- Ali MEK, Inanaga S, Sugimoto Y. Sources of resistance to Fusarium wilt of chickpea in Sudan. Phytopathol. Mediterr. 2002;41:163–169.
- Jendoubi W, Bouhadida M, Boukteb A, Béji M, Kharrat M. Fusarium wilt affecting chickpea crop. Agriculture. 2007;7:1-23
- Mohamed OE, Ahmed S, Singh M, Ahmed NE. Association of crop production practices on the incidence of wilt and root rot diseases of chickpea in the Sudan. Arab Journal of Plant Protection. 2018;36:21-26.
- Mohamed OE, Aladdin H, Seid A, Ahmed NE. Genetic variability of Fusarium oxysporum f. sp. ciceris population affecting chickpea in the Sudan. Journal of Phytopathology. 2015;163:1941-1946.
- Navas-Cortés JA, Landa BB, Mendez-Rodriguez MA, Jimenez-Díaz RM. Quantitative modeling of the effects of temperature and inoculum density of Fusarium oxysporum f. sp. ciceris races 0 and 5 on the development of Fusarium wilt in chickpea cultivars. Phytopathology. 2007;97:564-573.
- Iqbal MJ, Iftikhar K, Ilyas MB. Evaluation of chickpea germplasm for resistance against wilt disease. J Agri Res. 1993;31:449-453.
- Suliman W. Race identification of chickpea wilt pathogen (Fusarium oxysporum f. sp. ciceri) Hudeiba Research Station, Annual Report 2003/04,Sudan.
- Sheikh Mohamed AI. Chickpea improvement. In: Production and Improvement of Cool-Season Food Legumes in the Sudan (Salih SH, Ageeb OA, Saxena MC, Solh MB. eds), Proceedings of the National Research Review Workshop, Agricultural Research Corporation, Sudan/International Center for Agricultural Research in the Dry Areas, Syria/Directorate General for International Cooperation, The Netherlands. 1996;46-53.
- Mohamed AA, Tahir ISA, Elhashimi AMA. Assessment of genetic variability and yield stability in chickpea (Cicer arietinum L.) cultivars in River Nile State, Sudan. J Plant Breed Crop Sci. 2015;7:219-224.
- Mohamed AA, Ali SHN. Performance and stability of eight chickpea (Cicer arietinum L.) cultivars in different soil types at River Nile State, Sudan. Basic Res J Agirc Sci Rev. 2015;4:187-192.
- Ahmad MA, Iqbal SM, Najma A, Yasmin A, Abida A. Identification of resistant sources in chickpea against Fusarium wilt. Pak J Bot. 2010;42(1):417-426
- Chaudhry MA, Ilyas MB, Muhammad F, Ghazanfar MU. Sources of resistance in chickpea germplasm against fusarium wilt. Mycopath. 2007;5(1):17-21
- Chaudhry MA, Muhammad F, Afzal M. Screening chickpea germplasm against Fusarium wilt. J Agric Res. 2006;44(4):307-312.
- Halila MH, Strange RN. Screening of Kabuli chickpea germplasm for resistance to Fusarium wilt. Euphytica. 1997;96(2):273-279.
- Infantino A, Kharrat M, Riccioni L, Clarice J, Kevin E, Niklaus J. Screening techniques and sources of resistance to root diseases in cool season food legumes. Euphytica. 2006;147(1):201-221.
- Iqbal SM, Abdul G, Ahmad B, Iftikhar A, Altaf S. Identification of resistant sources for multiple disease resistance in chickpea. Pak J Phytopathol. 2010;22(2):89-94.
- Jiménez-Díaz RM, Crinó P, Halila MH, Mosconi C, Trapero-Casas A. Screening for resistance to Fusarium wilt and Ascochyta blight in chickpea. In: Breeding for stress tolerance in cool-season food legumes. Singh KB and Saxena MC, eds. John Wiley and Sons, Chichester, England, UK. 1993;77-96.
- Nen YL, Haware MP. Screening chickpea for resistance to wilt. Plant Dis. 1980;64:379-380.
- Sharma KD, Muehlbauer FJ. Fusarium wilt of chickpea: Physiological specialization, genetics of resistance and resistance gene tagging. Euphytica. 2007;157:1-14.
- Sharma KD, Chen W, Muehlbauer FJ. Genetics of chickpea resistance to five races of Fusarium wilt and a concise set of race differentials for Fusarium oxysporum f. sp. ciceris. Plant Dis. 2005;89:385-390.
Citation: Mohamed OE, Mohamed AE (2020) Screening of Chickpea (Cicer arietinum) Genotypes to Three Identified Races of Fusarium oxysporum f. sp. ciceris in Sudan. J Plant Pathol Microbiol 11: 507. doi: 10.35248/2157-7471.20.11.507
Copyright: © 2020 Mohamed OE, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

