Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2022) Volume 13, Issue 4
Role of Cow Dung on the Transmission of Enset Bacterial Wilt Pathogen (Xanthomonas campestris pv. musasearum) in Ethiopia
Misgana Mitiku1*, Alamar Seid1, Sadik Muzemil1, Zerihun Yemataw2 and Agdew Bekele32Department of Agriculture, Hawassa Agricultural Research Center, SARI, P.O.Box 1226, Hawassa, Ethiopia
3Department of Agriculture, Southern Agricultural Research Institute, SARI, P.O.Box 06, Hawassa, Ethiopia
Received: 20-Apr-2022, Manuscript No. JPPM-22-16207; Editor assigned: 25-Apr-2022, Pre QC No. JPPM-22-16207 (PQ); Reviewed: 09-May-2022, QC No. JPPM-22-16207; Revised: 16-May-2022, Manuscript No. JPPM-22-16207 (R); Published: 23-May-2022, DOI: 10.35248/2157-7471.22.13.608
Abstract
Enset (Ensete ventricosum) (Welw.) Cheesman is a monocarpic, herbaceous plant belonging to the Musacea family and the genus Ensete. All plant parts are utilized for different purposes. Hence, kocho, bulla and amicho are used as human food. By products from the plant is used to make different household items. Despite its importance, enset is constrained by several biotic and abiotic factors that affect its production and productivity. Among the biotic factors, diseases, insect pests and wild animals are important production challenges of enset. But, of all, Enset Bacterial Wilt (EBW) caused by Xanthomonas campestris pv. musacearum (Xcm) is the most damaging constraint contributing a major share to reduction of enset productivity in all enset growing areas of Ethiopia. Concerning sources of inoculum, mode of infection and transmission of Xcm especially of enset in Ethiopia is not yet well studied to the level needed as compared to the seriousness of the disease so, the current study was designed to evaluate the level of enset bacterial wilt pathogen transmission with animal excrete/cow dung. To do this a cow and calf allowed to feed only on the infected enset plant at morning, mid-day and night for seven consecutive days then in the morning and at evening dung and urine samples were collected for seven consecutive days from these animals and a total of 56 samples were collected. Then preparation of suspension and isolation of pathogen was made using standard bacterial isolation techniques. During the course of the experiment and counting of bacterial colony and biochemical test (KOH had been conducted for confirmation. The result of the experiment showed that from the total sample collected from cow and calf dung and urine only 19 or 33.92% were identified as Xanthomonas campestris pv. musacearum (Xcm) which was confirmed based on morphological and biochemical characteristics of the cultured isolates/samples. The rest 37 or 66.07% of the collected samples were didn’t show the specific characteristics of Xanthomonas campestris pv. musacearum (Xcm) so, they are not Xanthomonas campestris pv. musacearum (Xcm). From the total samples the highest average number of colony 19, 17.66, 14.66 and 13 were recorded from samples collected from calf dung at the morning, calf dung at the evening and cow dung at the morning respectively. Whereas the lowest average number of colony 4.66, 3, 2.33, 1.33, and 0.33 were recorded from samples collected from cow dung morning, cow dung morning, calf dung morning, cow urine morning and cow urine morning respectively. So from this study we can conclude that animal excrete both dung and urine have a role in transmitting the bacterial wilt of enset from one field to another at different level of transmission. Also the study depicted that large number of bacterial colony were isolated from dung than urine in the same way from calf than cow; so we should take care-off when we dispose the animal excrete and have to avoid the movement animals and disposing of the their waste materials in enset fields. Since, this is one time study, similar study has to be continued considering the age of animals and enset plant to have a clear understanding and information.
Keywords
Enset; XCM; Cow dung; Calf urine; Cow urine; Calf dung; Colony
Introduction
Enset (Ensete ventricosum) (Welw.) Cheesman is a monocarpic, herbaceous plant belonging to the Musacea family and the genus Ensete. Wild E. ventricosum is common and widespread in Ethiopia and along the rift valley in eastern Africa, all the way south to Mozambique. However, it is only in Ethiopia that enset has been domesticated and is cultivated for food, animal feed and fiber [1].
All plant parts are utilized for different purposes. Hence, kocho, bulla and amicho are used as human food. By products from the plant is used to make different household items. Fresh enset leaves are used as bread and food wrappers, cattle feed, serving plates, and pit liners to store kocho for fermentation. Though, the plant has multipurpose uses, it is principally cultivated for its carbohydrate rich food products. It is used as staple or co-staple food for about 20% of Ethiopian population particularly in the South and Southwestern part of the country.
Despite its importance, enset is constrained by several biotic and abiotic factors that affect its production and productivity. Among the biotic factors, diseases, insect pests and wild animals are important production challenges of enset. But, of all, enset bacterial wilt (EBW) caused by Xanthomonas campestris pv. musacearum (Xcm) is the most damaging constraint contributing a major share to reduction of enset productivity in all enset growing areas of Ethiopia [2].
A bacterium causing a wilt disease of enset was officially reported in Ethiopia for the first time in the 1960’s and named Xanthomonas musacearum. The same bacterium was later confirmed as causing a similar disease on cultivated banana and other Musa spp. and was subsequently reclassified as X. campestris pv. Musacearum.
Concerning sources of inoculum, mode of infection and transmission of Xcm especially of enset in Ethiopia is not yet well studied to the level needed as compared to the seriousness of the disease. However, a few studies have generally revealed some facts on sources of inoculum, mode of infection, transmission and survival of Xcm. Works done on dissemination mechanism of the pathogen have indicated to be mediated by several factors. Study on insect transmission of Xcm on enset suggested the possibility of transmission of the pathogen by insect vectors [3].
Farmers also claim that the pathogen is transmitted by cow dung when the cows eat infected enset. However, this fact is not yet fully clarified so, the current study was designed to evaluate the level of enset bacterial wilt pathogen transmission with animal excrete/ cow dung
Materials and Methods
Feeding of diseased enset plant to the animal and collection of dung and urine samples
Diseased enset plant was collected from the farmer’s field found at Woyibo kebeles of Damot sore woreda of Wolaita zone. This area was selected for diseased enset plant collection as it represents the major of enset production areas in zone [4]. Diseased enset plant was transported to the area where the animals were kept isolated from their normal feeding habit for this purpose; then these animals were allowed to feed only on the infected enset plant at morning, mid-day and night for seven consecutive days then in the morning and at evening dung and urine samples were collected for seven (7) consecutive days from these animals feeding on the infected enset plant so, a total of 56 samples were collected.
Preparation of suspension and pathogen isolation
Preparation of suspension for pathogen isolation was conducted using the collected urine and dung samples. To make the suspension 100 ml of distilled water was added in each collected samples and poured using 15 m sieve to separate the course materials from the suspension and the collected suspension was transferred to a new jar then preparation of the working dilution (10-1 to 10-4) was made following the standard dilution techniques; then after isolation of the pathogen was carried out using artificial growth media. (Figure 1).
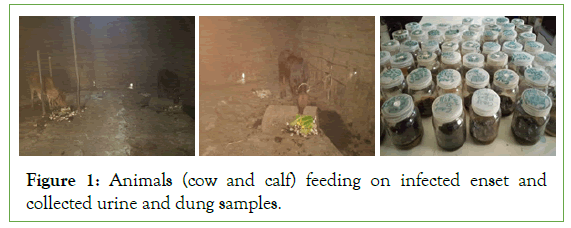
Figure 1: Animals (cow and calf) feeding on infected enset and collected urine and dung samples.
Before isolation of the pathogen 1 litter of Yeast Dextrose Calcium Carbonate Agar (YDCA) growth media was prepared using 20 g of dextrose, 10 gram of yeast extract, 20 g of calcium carbonate,10 ml of glycerol,15 gram of agar-agar instead of bacteria agar and 990 ml of distilled water [5,6].
To prepare it first we mixed the entire ingredient together gently and autoclaved at 121°C for 15 min then upon cooling we poured it on to Petri plate inside the hood and we made ready it for growing of pathogen. For isolation of the pathogen a loop-full of suspension from the sample were spread on to Yeast Dextrose Calcium Carbonate Agar (YDCA) growth medium using inoculation loop, and then incubated at 28°C for 24–48hrs. After 48hrs, colonies of the bacterium were observed. Pure culture of the bacteria was prepared by transferring individual colony of the bacterium on to YDCA culture medium and incubated at 28°C for 24 to 48 h. (Figure 2).

Figure 2: Preparation of suspension, growth media and culturing of pathogen.
Biochemical test
KOH Test: Two drops, approximately 50l, of a 3% (w/v) solution of Potassium Hydroxide (KOH) were placed on a clean glass slide. Bacterial cells were transferred from culture media aseptically with a flat wooden toothpick and placed into the drop of KOH with rapid circular agitation. After 5-8 sec, the toothpick was alternately raised and lowered just off the slide surface to detect with a stringing effect. The KOH test was considered positive if, drop viscosity increased and stringing occurred within 15 sec. (Figure 3).
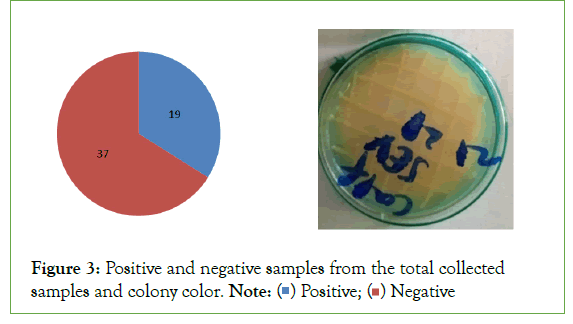
Figure 3:Positive and negative samples from the total collected samples and colony color.
Data collection and analysis
Number of colony grown on the growth medium, growth characteristics on growth media and biochemical reaction of the pathogen were noted and analysed using simple statistics [7].
Results and Discussion
Morphological and Colony number
The isolation result show that from the total sample collected from cow and calf dung and urine only 19 or 33.92% were identified as Xanthomonas campestris pv. musacearum (Xcm) (Figure3) which was confirmed based on morphological and biochemical characteristics of the cultured isolates/samples. The rest 37% or 66.07% of the collected samples were didn’t show the specific characteristics of Xanthomonas campestris pv. musacearum (Xcm) so, they are not Xanthomonas campestris pv. musacearum (Xcm).
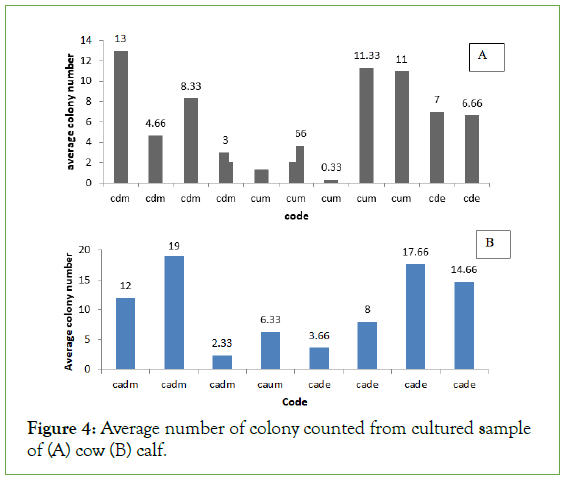
From the total collected samples the highest average number of colony 19, 17.66, 14.66 and 13 were recorded from samples collected from calf dung at the morning, calf dung at the evening and cow dung at the morning respectively. Whereas the lowest average number of colony 4.66, 3, 2.33, 1.33, and 0.33 were recorded from samples collected from cow dung morning, cow dung morning, calf dung morning, cow urine morning and cow urine morning respectively (Figures 4a and 4b).
Figure 4: Average number of colony counted from cultured sample of (A) cow (B) calf.
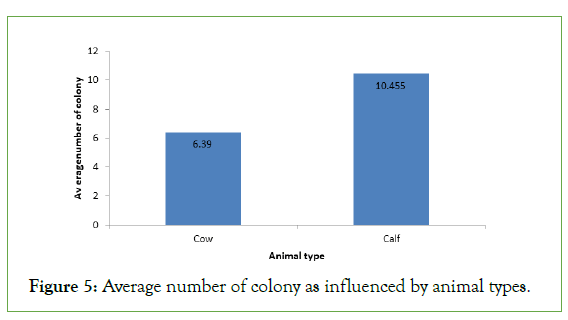
Regarding average number of colony isolated from different types of animals, the highest average number of colony 10.455 was recorded samples collected from calf this may be due to the high feed intake and digestion capacity of the animal and also the age of the animal (Figure 5).
Figure 5: Average number of colony as influenced by animal types.
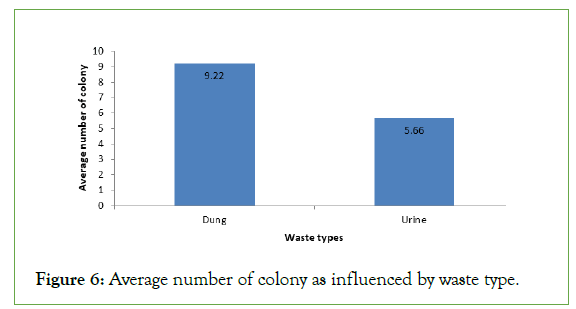
The results also show that the type of animal waste also gave different average number of colony. The highest average number of colony 9.22 was counted from the dung sample of the animals whereas the lowest average number of colony was recorded from urine sample of the animals (Figure 6). This implies that the dung can support much number of bacterial colonies than urine (Figure 7).
Figure 6: Average number of colony as influenced by waste type.
Figure 7: dead leaves of inoculated enset plant.
Biochemical test result
Xanthomonas campestris pv. musacearum (Xcm) samples used for biochemical test were selected based upon having typical yellow colony morphology and being the intended bacteria isolated from these samples. Of these isolates, 68.42% were isolated from dung samples whereas 31.57% were from urine samples [8].
The biochemical tests showed positive reaction for KOH test for 19 samples. So, biochemical properties of 33.92% of Enset Bacterial Wilt (EBW) isolates generally were consistent with that of Xanthomonas campestris pv. musacearum (Xcm) (Table1).
| XCM sample code | Biochemical Tests | |
|---|---|---|
| KOH (3%) | Designation | |
| cdm | + | Gm- |
| cdm | + | Gm- |
| cum | + | Gm- |
| cade | + | Gm- |
| cde | + | Gm- |
| caum | + | Gm- |
| cdm | + | Gm- |
| cum | + | Gm- |
| cde | + | Gm- |
| cadm | + | Gm- |
| cum | + | Gm- |
| cade | + | Gm- |
| cadm | + | Gm- |
| cdm | + | Gm- |
| cum | + | Gm- |
| cade | + | Gm- |
| cadm | + | Gm- |
| cum | + | Gm- |
| cade | + | Gm- |
Note: cdm=cow dung morning, cum=cow urine morning, cade=calf dung evening, cde=cow dung evening, caum=calf urine morning, cadm=calf dung morning.
Table 1: Biochemical reaction for identified XCM pathogen from dungb and urine samples of cow and calf.
Pathogencity test for confirmation of the pathogen
For the pathogenecity test one year old susceptible enset clone (Arkya) was planted on pot then inoculated with 5 ml of a 2 days old bacterial suspension with a cell concentration of 108 cfu/ml (adjusted to 0.3 OD at 460 nm using spectrophotometer). The plant was inoculated at the base of the newly expanding central leaf petiole using a 10ml capacity sterile hypodermic syringe with metal needle. Then symptom was seen after 21 days after inoculation and inoculated leaves were dead after 45 days of inoculation.
Conclusion and Recommendation
This study was conducted in Areka Research Center in laboratory condition with the aim of two evaluating the level of enset bacterial wilt pathogen transmission with animal excrete/cow dung. A total of 56 dung and urine sample was collected from two types of animals (Cow and calf). So from this study we can conclude that animal excrete both dung and urine have a role in transmitting the bacterial wilt of enset from one field to another at different level of transmission. Also the study depicted that large number of bacterial colony were isolated from dung than urine in the same way from calf than cow; so we should take care-off when we dispose the animal excrete and have to avoid the movement animals and disposing of the their waste materials in enset fields. Since, this is one time study, similar study has to be continued considering the age of animals and enset plant to have a clear understanding and information.
Acknowledgement
The study was financed by the McKnight Foundation Collaborative Crop Research Program Project, “Integrated Management of Bacterial Wilt of Enset (Ensete ventricosum) (Welw.) Cheesman caused by Xanthomonas campestris pv. musacearum in Ethiopia’’. We thank Areka, Agricultural Research Center and Southern Agricultural Research Institute for hosting and provision of necessary services and facilities during field and laboratory work.
REFERENCES
- Bezuneh T, Feleke A, Bayie R. The cultivation of the genus Ensete in Ethiopia. InSoil Crop Sci Soc Fla Proc. 1967;27:133-141).
- Spring A, Diro M, A Brandt S, Tabogie E, Wolde-Michael G, McCabe JT, et al. Tree against hunger: enset-based agricultural systems in Ethiopia. AAAS. 1997; 55.
[Crossref], [Google scholar]
- Handoro F, Said A. Enset clones responses to bacterial wilt disease (Xanthomonas campestris pv.Musacearum). IJAPSA. 2016;2(09): 45-53.
- Wolde M, Ayalew A, Chala A. Assessment of bacterial wilt (Xanthomonas campestris pv. musacearum) of enset in Southern Ethiopia. Afr. J. Agric. Res. 2016; 1(19): 1724-1733.
[Crossref], [Google scholar]
- Tensaye AW, Lindén B, Ohlander L. Enset farming in Ethiopia. I. Soil nutrient status in Shoa and Sidamo regions. Commun Soil Sci Plant Anal. 1998;29(1-2): 193-210.
[Crossref], [Google scholar]
- Wondimagegn, E. 1981. The role of Poecilocarda nigrinervis, Pentalonia nigeronervosa and Planococus ficus in the Transmission of Ensete wilt Pathogen, Wolaita, Ethiopia, MSc. thesis. Addis Ababa University.
- Yirgou D, Bradbury JF. A note on wilt of banana caused by the enset wilt organism Xanthomonasmusacearum. East Afr agric for j. 1974;40(1): 111-114.
[Crossref], [Google scholar]
- Young JM, Dye DW, Bradbury JF, Panagopoulos CG, Robbs CF. A proposed nomenclature and classification for plant pathogenic bacteria. N Z J Agric Res. 1978;21(1): 153-177.
[Crossref], [Google scholar]
Citation: Mitiku M, Seid A, Muzemil S, Yemataw Z, Bekele A (2022) Role of Cow Dung on the Transmission of Ensete Bacterial Wilt Pathogen (Xanthomonas campestris pv. musasearum) in Ethiopia. J Plant Pathol Microbiol. 13:608.
Copyright: © 2022 Mitiku M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.