Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 14, Issue 6
Reverse Transcriptase Loop-Mediated Isothermal Amplification Assays (RTLAMP) for the Detection of Alfalfa Mosaic Virus from Forage Crops
Woubit Dawit*, Fikerte Mulatu, Yesuf Eshete and Alice MuchugiFeed and Forage development program, International Livestock Research Institute (ILRI), Nairobi, Kenya
Received: 21-Jul-2023, Manuscript No. JPPM-23-22301; Editor assigned: 24-Jul-2023, Pre QC No. JPPM-23-22301 (PQ); Reviewed: 07-Aug-2023, QC No. JPPM-23-22301; Revised: 22-Aug-2023, Manuscript No. JPPM-23-22301 (R); Published: 29-Aug-2023, DOI: 10.35248/2157-7471.23.14.690
Abstract
AMV is one of the most economically important plant viruses and has a very wide host range including forage crops. To make sure that only seeds of satisfactory phytosanitary status are distributed to recipients in geographically diverse areas, the ILRI genebank routinely monitors seed-borne diseases during seed multiplication and in the field genebank. Current detection techniques for AMV include a dot-blot assay and a two-step Reverse Transcription Polymerase Chain Reaction (RT-PCR), both of which are time consuming. In the present study we developed a onestep Reverse Transcriptase Loop-Mediated Isothermal Amplification (RT-LAMP) method for the sensitive and rapid detection of AMV from forage crops. For the assay, six different AMV specific primers were used, and a series of reactions were performed to identify optimal conditions. Amplicons were easily visualized by means of an in-tube colour indicator (SYBR Green I dye) with no requirement to run gel electrophoresis. The sensitivity of the RT-LAMP assay was assessed by comparing the optimized AMV RT-LAMP assay with the conventional RT-PCR. In RT-LAMP, an amplicon was generated up to 100 ag/μl dilutions in contrast to the conventional RT-PCR, where no amplification at 1 fg/μl and onwards, indicating that the detection limit of the AMV RT-LAMP assay is much lower than for the conventional RT-PCR. Finally, the optimized RT-LAMP assay was further validated on 40 field samples of different forage species and other important forage viruses. The developed RT-LAMP assay was specific and detected AMV from different forages with no cross reaction with other plant viruses (SBMV and potyvirus) tested in the ILRI-genebank seed multiplication/regeneration fields. The optimized RT-LAMP assay is an effective tool for the detection of AMV for field samples in diagnostic laboratories, and for quarantine applications.
Keywords
Forage; AMV; RT-LAMP; RT-PCR; Dot-blot
Introduction
Alfalfa Mosaic Virus (AMV) is a pathogen that causes a widespread disease that results in yield losses of up to 41% for shoot dry weight and 45% for seeds [1]. AMV was first isolated from alfalfa (Medicago sativa), which is the main natural host, and the pathogen infects over 165 species, mainly in the Fabaceae and Solanaceae families [2]. The virus is seed-borne and can also be transmitted by several aphids and by mechanical inoculation [3]. AMV isolates are classified into distinct strains and variants [4,5] which result in the virus having an extended host range and makes it one of the most economically important plant diseases worldwide [6,7]. The International Livestock Research Institute (ILRI) maintains over 18,664 accessions of grass and legume forages in trust in its forage genebank with the aim of making seeds freely available for evaluation and subsequent incorporation into sustainable crop-livestock systems. To make sure that only seeds of satisfactory phytosanitary status are distributed to recipients in geographically diverse areas, the ILRI genebank routinely monitors seed-borne diseases during seed multiplication and in the field genebank . Mih and Hanson detected AMV in 24 leguminous forage species conserved at the ILRI genebank, of which 17 species were reported for the first time as a natural host for AMV [8].
Detection methods for AMV predominantly focus on RT-PCR and serological assays [9]. However, these protocols either require a long reaction time, multiple steps, sophisticated machines, and specialists to run the experiments. Moreover, use of PCR for field level surveys and for routine screens of large samples has been limited largely by the high costs and requirements for well-established laboratories [10]. Development of a time and cost-efficient method for virus detection with high efficiency and specificity is essential to perform field studies and for cleaning of infected germplasm. Loop-Mediated Isothermal Amplification (LAMP) is a molecular technique developed as an alternative to PCR-based technologies [11]. It is a one-step amplification of target DNA/RNA at a single temperature. It is a highly efficient and fast protocol that is specific to the target sequence because of the use of four or six primers targeting six or eight different regions, respectively [12,13]. The LAMP assay has been used for the molecular detection and diagnosis of many pathogens, including bacteria, viruses, fungi, and parasites responsible for plant, animal, and human diseases [14,15]. RT-LAMP has been developed to detect several plant viruses, including members of the genera Potyvirus [16-19]. In this study, we developed RT-LAMP for rapid and specific AMV detection from forage crops and the sensitivities of RT-LAMP was compared with the standard RT-PCR assay.
Materials and Methods
Reference sample and investigated virus samples
In 2022 thirty (30) alfalfa leaf samples that showed symptoms typical of AMV, mottling and mosaic symptoms, were collected from the ILRI regeneration field/greenhouse, Addis Ababa, Ethiopia. The samples were assayed for AMV infection using a combination of dot-blot assay, and RT-PCR following the procedure described below. These known positive samples were used for optimization of the RT-LAMP protocol. Forty (40) leaf samples of Alfalfa, Urochloa spp., Neonotania wightii and Trifolium spp., collected from the regeneration field and greenhouse were used to validate the optimized RT-LAMP assay. In addition, pathogens causing the major viral diseases of forages, Southern Bean Mosaic Virus (SBMV) and Potyvirus, were maintained/included to validate the optimized RT-LAMP assay.
Dot-blot assay
The dot-blot assay was carried out using the method described by Cardosa et al. [20]. Briefly, the crude extracts (6 μl) from homogenized leaves extracted in carbonate buffer comprising 2% Polyvinyl Pyrrolidine (PVP, MW 40,000) were spotted onto a pre-washed Nitro Cellulose Membrane (NCM), soaked in 1x Phosphate Buffer Saline (PBS), pH 7.2. The NCM was air-dried, and the free binding sites were blocked by soaking the membrane in 5% non-fat skimmed milk prepared in 1x PBS for 30 minutes. All the membranes coated with the samples were then washed five times at 10-minute intervals with 1x PBS, pH 7.2. Then, the membranes coated with the sample extracts were soaked in specific antiserum (DSMZ, Germany) diluted (1:1000 dilution) in PBS-TPO (PBS with 0.05% Tweenty 20, 2% PVP and 0.2% ovalbumin) and incubated at 37°C for 1 hour (hr), followed by incubation for another 1 hr at 37°C in a 1:3000 dilution of goat anti-rabbit IgG-Alkaline phosphatase conjugate (Sigma Aldrich, St. Louis, USA) prepared in PBS-TPO. Subsequently, the membranes were incubated in freshly prepared substrate solution, 5-Bromo-4-Chloro-3-Indolyl Phosphate (BCIP)/4-Nitro Blue Tetrazolium chloride (NBT) in dark conditions, by covering the tray with an Aluminum sheet, at 37°C for 15-20 mins or until the development of a purple blue colour. All the membranes were washed with 1x PBS for 5-times at 10 minutes intervals after each step of the dot-blot assay.
Reverse Transcriptase PCR (RT-PCR)
RNA extraction: Total RNA was extracted from freeze dried leaf samples using the TRI Reagent® protocol [21]. As a reference, RNA was also extracted from the freeze dried inocula of AMV and SBMV, obtained from the German collection of microorganisms and cell cultures (DSMZ), Germany. Briefly, approximately 50 mg of freeze-dried leaf material was homogenized using a mortar and pestle and mixed with 1 ml of Trizol reagent (Thermo Fisher Scientific, USA) followed by phase separation by extracting with an equal volume of chloroform, and centrifugation (12,000 × g for 15 mins at 4°C). The aqueous phase was pipetted into a clean tube, the RNA was precipitated using isopropyl alcohol, washed with 75% ethanol, and re-dissolved in RNase-free water. The quality and quantity of RNA was determined using a nanodrop spectrophotometer (DeNovix DS-11, Thermo Fisher Scientific).
RT-PCR amplification: A pair of AMV specific primers, which were designed from the Coat Protein (CP) region of the virus at the ILRI-Genebank (Unpublished) were used for detection of AMV in the samples. The primers include, AMV: GBV53F-5’GGATCCATGAGTTCTTCACAAAAGAAAGCT-3’ and GBV97R: 5’AGCCCCACAGTAATCAAACTG-3’. The virus was detected by RT-PCR in a two-step process in which the cDNA was first synthesized in a 20 μl reaction mix, where 2 μl of RNA template (~500 ng) was mixed with 1 μl each of OligodT primer and 10mM dNTP mix with the volume adjusted to 15 μl with nuclease free water. The mixture was heated to 65°C in a thermocycler for 5 mins followed by addition of 4μl of 5X RT buffer and 1 μl Maxima H Minas Enzyme mix (ThermoFisher, Scientific, USA). The first strand cDNA synthesis was conducted by incubating the mixture at 50°C for 30 mins followed by 85°C for 5 minutes. Then, a PCR was carried out in a 25 μl reaction volume containing 2 μl of cDNA, 12 μl DreamTaq Master Mix (Thermo Fisher, USA), 1 μl of 10 μM each forward and reverse primer, and 9 μl of nuclease free water. The thermal cycling conditions were an initial denaturation step at 98°C for 30 s followed by 35 cycles of 98°C for 10 s, annealing temperature of 54°C for 1 min, and 72°C for 1 min with a final extension at 72°C for 10 min. The amplified products were resolved by electrophoresis on a 1.5% agarose gel and visualized under a UV gel documentation system (Intas, Germany).
RT-LAMP primer and assay optimization
LAMP primers: AMV specific LAMP primers, first reported by Almasi and Almasi [22], designed from conserved sequence of the CP gene were used. These include forward outer F3 (5’-TTTAAACCTTGATCATTTGCTGGA-3’), reverse outer B3 (5’-ATGGGTTTTAGAGCATATTCTACT-3’), forward inner FIP (5’- GTCATTCTTAACCCCGTCGTTTTTT -3’), reverse inner BIP (5’- TGCAATTAATTCTTAACGGATTTTT-3’), loop forward Loop LF (5’- AT CGAACACACGTGCAACCC-3), and loop reverse Loop, LB (5’- TCC ATTTATATTCGGGGATGT-3’). Primers were synthesized by Invitrogen™, Macrogen, Korea.
Assay optimization: A series of reactions were performed using the AMV primers to obtain optimal conditions. Key considerations in the development and optimization of LAMP assays were the effect of different concentrations of dNTPs, Bst Tag polymerase, and reverse transcriptase. For dNTPs, the final concentration of 0.2 to 10 mM and for Bst DNA polymerase from 0 to 12 U were prepared. For the reverse transcriptase optimization 0 μl (without), 0.25 μl (50 U) and 0.5 μl (100 U) were used. LAMP reaction temperature (evaluated at 56-70°C) and incubation time 30, 40, 50, 60, 70, 80, 90, 100, and 110 min were also considered. The optimization was repeated three times as recommended by Liu et al. [23] and Warghane et al. [24]. When the isothermal amplification was completed, 5 μl of 100X SYBR Green I dye (Thermo Fisher scientific, USA) was added, and amplicons were evaluated by eye, as well as under ultraviolet light, for colour development. The amplicons were also separated by electrophoresis on a 1.5% agarose gel for further confirmation.
RT-LAMP reaction: 1 μl of 100 ng RNA was used as a template in the RT-LAMP reaction. The LAMP mixture contained the optimized concentrations: 2.5 μl 10X DNA polymerase buffer, 100 U reverse transcriptase, 2.5 μl 50 mM dNTPs mix, 2.0 μl 10 mM MgCL2, 0.5 μl 10 mM F3, 0.5 μl 10 mM B3, 2.0 μl 10 mM FIP, 2.0 μl 10 mM BIP, 1.0 μl 10 mM FL, 1.0 μl 10 mM BL, 1.0 μl 5 M Betaine (Sigma-Aldrich, USA), and 1.0 μl 8 U of Bst DNA polymerase, exonuclease Minus 2,000 U at 8U/μl (BIOSEARCH Technologies, UK). The mixture was incubated at 64°C for 60 min in a water bath. Finally, the amplification was analyzed by adding SYBR Green I dye and separating by gel electrophoresis.
Sensitivity of the RT-LAMP: The sensitivity of the RT-LAMP assay was assessed by comparing the optimized AMV RT-LAMP assay with the conventional RT-PCR. For this, AMV-RNA adjusted to 100 ng/μl was further serially diluted (10-fold serial dilutions) in a solution of total RNA isolated from a healthy alfalfa plant by taking 1 μl of serially diluted templates. A total of twelve (12) dilutions, starting from 100 ng/μl, to 1 ag/μl, was tested [25]. The reaction was performed with each dilution used as a template. The extracted total RNA from a healthy plant was used as a negative control.
Validation of RT-LAMP: the AMV RT-LAMP assay was validated by testing 40 samples of alfalfa, Neonotania wightii, Trifolium spp., and Urochloa spp., that were randomly taken from seed multiplication/regeneration field and two samples of RNA of each known positive sample of other important forage viruses, Potyvirus and SBMV. As a negative control, extracted total RNA from a healthy plant of the different forage species was used.
Results
Dot-blot assay and RT-PCR reaction
The dot-blot assay result revealed a strong to very weak purple colour on 4 out of 15 (27%) symptomatic alfalfa leaf samples obtained from the ILRI greenhouse. Whereas the RT-PCR successfully detected 14 (47%) of the samples infected by AMV, and the expected fragment (632 bp) was observed on agarose gel (Figure 1). This indicates that RT-PCR is more sensitive than the dot-blot assay. All the positive samples detected by the dot-blot were also detected by the RT-PCR. The positive samples were used for further LAMP assay optimization.
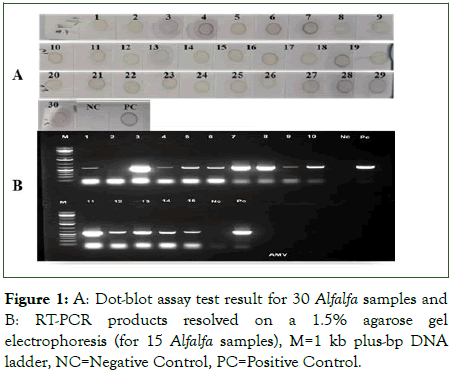
Figure 1: A: Dot-blot assay test result for 30 Alfalfa samples and B: RT-PCR products resolved on a 1.5% agarose gel electrophoresis (for 15 Alfalfa samples), M=1 kb plus-bp DNA ladder, NC=Negative Control, PC=Positive Control.
Optimized conditions and specificity of RT-LAMP reactions
The RT-LAMP assay was performed at different temperatures (56°C-70°C) and time durations (30-110 min) with total RNA from plant samples infected with AMV as a template for RT-PCR. Amplification results were assessed visually and under UV by colour changes, from orange to green, in samples incubated in a temperature between 58°C to 66°C, with optimum isothermal amplification of 64°C but not below 56°C and above 66°C as well as in the healthy negative control and the reaction control without template RNA (Figure 2). The same samples exhibited also ladder-like characteristics of binding pattern on agarose gel electrophoresis. Moreover, the analysis of the amplicon from different time duration indicated that the minimum time for completion of the reaction was 30 min, and the strongest colour intensity (amplification) was observed in a reaction that was performed at 60 min and onwards (Figure 3). To select the optimum concentration of dNTPs, Bst DNA polymerase and reverse transcriptase for the RT-LAMP reaction, tests were conducted in different concentration mixtures. The test results showed that at 5 mM and 10 mM dNTPs, ladder like DNA fragments were clearly observed (data not shown). For Bst DNA polymerase, even though amplification was observed/detected in all units of enzyme, the optimum amplification was observed at 6 U and 8 U (Figure 4). Similarly, optimum amplification was observed when reverse transcriptase was used at 0.5 μl (100 U). No amplified product was obtained when Bst DNA polymerase and reverse transcriptase were not added to the reaction mix.
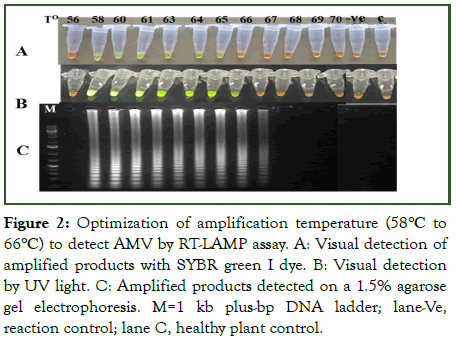
Figure 2: Optimization of amplification temperature (58°C to 66°C) to detect AMV by RT-LAMP assay. A: Visual detection of amplified products with SYBR green I dye. B: Visual detection by UV light. C: Amplified products detected on a 1.5% agarose gel electrophoresis. M=1 kb plus-bp DNA ladder; lane-Ve, reaction control; lane C, healthy plant control.
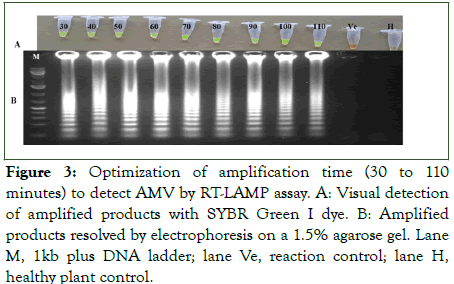
Figure 3: Optimization of amplification time (30 to 110 minutes) to detect AMV by RT-LAMP assay. A: Visual detection of amplified products with SYBR Green I dye. B: Amplified products resolved by electrophoresis on a 1.5% agarose gel. Lane M, 1kb plus DNA ladder; lane Ve, reaction control; lane H, healthy plant control.
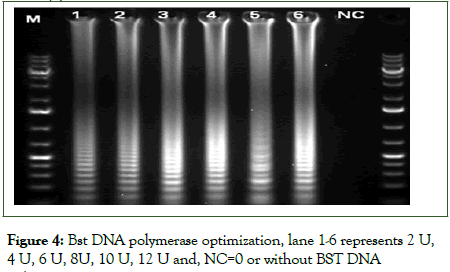
Figure 4: Bst DNA polymerase optimization, lane 1-6 represents 2 U, 4 U, 6 U, 8U, 10 U, 12 U and, NC=0 or without BST DNA polymerase.
Sensitivity of RT-LAMP and validation of LAMP assays using field samples
To compare the sensitivity of the RT-LAMP assay and the conventional RT-PCR, a 10-fold serial dilution of AMV RNA was used for the amplification reaction. In RT-LAMP, an amplicon was generated in up to 100 ag/μl dilutions but with decreased band intensity as dilution increased (Figure 5). In the conventional PCR, there was no amplification at 1 fg/μl and onwards.
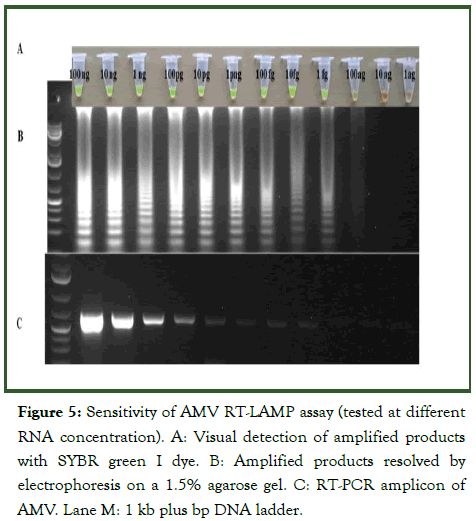
Figure 5: Sensitivity of AMV RT-LAMP assay (tested at different RNA concentration). A: Visual detection of amplified products with SYBR green I dye. B: Amplified products resolved by electrophoresis on a 1.5% agarose gel. C: RT-PCR amplicon of AMV. Lane M: 1 kb plus bp DNA ladder.
The optimized RT-LAMP assay was further validated on 40 symptomatic and asymptomatic field samples of different forage species. Out of the 40 samples, 33 samples were found to be positive for AMV when assayed with the RT-LAMP, and the same results were obtained by RT-PCR except for three samples (Table 1), which tested negative in RT-PCR. The results were consistent even when both assays were repeated in triplicate. Additionally, the assay did not show any cross-reactivity with other major forage virus pathogens (SBMV and Potyvirus), which showed the specificity of the protocol detecting only the targeted virus, AMV.
| S. no. | Accession no. | Type/species | Location | Detection by RT-LAMP | Detection by RT-PCR | |
|---|---|---|---|---|---|---|
| SYBR green I dye | Agarose gel | |||||
| 1 | SF730ql | Medicago sativa | Shola | + | + | + |
| 2 | SF730ql | Medicago sativa | Shola | + | + | - |
| 3 | SF730ql | Medicago sativa | Shola | + | + | + |
| 4 | SF730ql | Medicago sativa | Shola | + | + | + |
| 5 | SF730ql | Medicago sativa | Shola | + | + | + |
| 6 | Stamina GTG | Medicago sativa | Shola | + | + | + |
| 7 | Stamina GTG | Medicago sativa | Shola | + | + | + |
| 8 | Stamina GTG | Medicago sativa | Shola | + | + | + |
| 9 | Stamina GTG | Medicago sativa | Shola | + | + | + |
| 10 | Stamina GTG | Medicago sativa | Shola | + | + | + |
| 11 | DZ-ZMSFG100 gm | Medicago sativa | Shola | + | + | + |
| 12 | DZ-ZMSFG100 gm | Medicago sativa | Shola | + | + | + |
| 13 | Sardi grazer | Medicago sativa | Shola | + | + | + |
| 14 | Sardi grazer | Medicago sativa | Shola | + | + | + |
| 15 | Sardi grazer | Medicago sativa | Shola | + | + | + |
| 16 | 14755 | Urochloa brizantha | Ziway | + | + | + |
| 17 | 11042 | Urochloa brizantha | Ziway | + | + | + |
| 18 | 13499 | Urochloa brizantha | Ziway | + | + | + |
| 19 | 13777 | Urochloa brizantha | Ziway | + | + | + |
| 20 | 13726 | Urochloa brizantha | Ziway | + | + | - |
| 21 | 13462 | Urochloa brizantha | Ziway | + | + | - |
| 22 | 13819 | Urochloa brizantha | Ziway | + | + | + |
| 23 | 13363 | Urochloa brizantha | Ziway | + | + | - |
| 24 | 13598 | Urochloa brizantha | Ziway | + | + | + |
| 25 | 13799 | Urochloa brizantha | Ziway | - | - | - |
| 26 | 718-11 | Urochloa humidicola | Ziway | + | + | + |
| 27 | 735-3 | Urochloa humidicola | Ziway | + | + | + |
| 28 | 822-10 | Urochloa humidicola | Ziway | + | + | + |
| 29 | 832-5 | Urochloa humidicola | Ziway | + | + | + |
| 30 | 11022-6 | Urochloa humidicola | Ziway | + | + | + |
| 31 | 800-2 | Trifolium tembense | Shola | + | + | + |
| 32 | 6261-6 | Trifolium steudneri | Shola | - | - | - |
| 33 | 6777-3 | Trifolium tembense | Shola | + | + | + |
| 34 | 22264-1 | Neonotonia wightii | Shola | - | - | - |
| 35 | 22276-1 | Neonotonia wightii | Shola | + | + | + |
| 36 | 13071-4 | Neonotonia wightii | Shola | + | + | + |
| 37 | Inocula | SBMV +ve sample | DSMZ | - | - | - |
| 38 | Inocula | SBMV +ve sample | DSMZ | - | - | - |
| 39 | Inocula | Potyvirus +ve sample | DSMZ | - | - | - |
| 40 | Inocula | Potyvirus +ve sample | DSMZ | - | - | - |
Table 1: Detection of AMV in symptomatic and asymptomatic samples and samples infected by other viruses using Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) and Reverse Transcription Polymerase Chain Reaction (RT-PCR).
Discussion
AMV is one of the most important seed-borne plant viruses occurring worldwide and it naturally infects over 150 herbaceous and woody species in 22 plant families. In Ethiopia, the virus has been reported in several forage species and 17 species were reported for the first time in world literature. Previous detection methods for AMV focused predominantly on RT-PCR and dot-blot assays. However, these two protocols require a long reaction time, multiple steps, expensive machines, and specialists to run the test. Development of a time and cost-efficient method for virus detection with high efficiency and specificity is essential to perform field studies and for cleaning of infected germplasm of quarantine pathogens at the ILRI-genebank. In the present study we optimized a one-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) method for the sensitive and rapid detection of AMV from forage crops. The developed RT-LAMP assay in the present study is specific and effectively detected AMV from different forage crops while not cross reacting with other important forage viruses (SBMV and Potyvirus).
The dot-blot assay, as compared with RT-PCR and RT-LAMP, commonly takes a long time to complete the test steps from extraction to detection. In addition, the assay is less sensitive to detect positive samples particularly from asymptomatic plants which may be related to the inoculum concentration/amount in the sample. Garnsey [26] and Sharma et al., [27] reported that serological methods that use antisera for detection are less sensitive than techniques that target nucleic acid. Moreover, it is often limited by the need to have a continuous supply of good quality antisera and lacks resolution when virus strains are closely related.
RT-PCR is a very sensitive and more widely used technique than the serological techniques. In the present study out of 30 Alfalfa samples tested, 8 (27%) and 14 (47%) were found to be positive with the dot-blot assay and RT-PCR, respectively. However, the RNA extraction and cDNA production takes a long time (more than 4 hrs. in our case) for the RT-PCR assay. In the RT-LAMP procedure, the extracted RNA is used directly as a template and separation of the amplicons by gel electrophoresis is not required. Hence, RT-LAMP takes less time to assay for the pathogen compared to RT-PCR. In the present study, the overall time required to accomplish the test for RT-LAMP was 60 min. The amplified products can be easily visualized by means of in-tube colour indicator (SYBR green I dye) with no essential requirement for additional staining systems and gel electrophoresis.
The specificity and sensitivity of the RT-LAMP assay were further validated through comparison with conventional RT-PCR. When a 10-fold serial dilution of RNA was used as a template, an amplicon was generated in dilutions of up to 100 ag/μl in the case of RT-LAMP, but no amplification at 1 fg/μl and onwards was observed in the case of RT-PCR, this indicates that RT-LAMP assay is more sensitive than the conventional RT-PCR. In addition, out of the 40 field samples tested, 33 for RT-LAMP and 30 for RT-PCR samples were found to be positive. The result was consistence even the three samples were tested in several replicates. This discrepancy may have arisen because the concentration of the virus RNA in these three (3) samples was very low and beyond the detection limit of the RT-PCR assay. Ferna´ndez-Soto et al. [13] reported that, the sensitivity of the RT-LAMP assay is higher than that of RT-PCR; consequently, the RT-LAMP assay could successfully detect the virus even at low concentration. The LAMP assay has been used for the detection and diagnosis of many pathogens, including bacteria, viruses, fungi, and parasites responsible for plant, animal, and human diseases. The present study demonstrated the use of the LAMP protocol for the detection of AMV virus from diverse forage species. Therefore, we recommend the optimized RT-LAMP for effective and rapid detection of AMV from field samples in diagnostic laboratories, and for quarantine applications.
Conclusion
RT-LAMP assay was successfully developed for rapid, specific, and sensitive detection of AMV from different forages with no cross reaction with other forage viruses. The assay was more sensitive compared to RT-PCR. In addition, amplicons were easily visualized by means of an in-tube colour indicator (SYBR Green I dye) with no requirement to run gel electrophoresis. Therefore, the optimized RT-LAMP assay is an effective tool for the detection of AMV for field samples in diagnostic laboratories, and for quarantine applications.
Acknowledgment
This study was conducted with financial support from CGIAR Genebank Initiative aims to strengthen the global genebank system in conserving and making available a wide variety of plant genetic resources.
Conflicts of Interest
The authors have no conflict of interest to declare that are relevant to this article.
Funding
The research was funded by CGIAR Genebank initiative.
Financial Interests
All authors declare they have no financial interests.
Author Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Woubit Dawit, Fikerte Mulatu, and Yesuf Eshete. The first draft of the manuscript was written by Woubit Dawit, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
References
- Latham LJ, Jones RAC, Coutts BA. Yield losses caused by virus infection in four combinations of non-persistently aphid-transmitted virus and cool-season crop legume. Australian J Exp Agricul. 2004;44:57-63.
- He B, Fajolu OL, Wen RH, Hajimorad MR. Seed transmissibility of Alfalfa mosaic virus in soybean. Plant Health Prog. 2010;11(41):1-5.
- Jaspars EM, Bos L. Alfalfa mosaic virus. No. 229 in: Descriptions of plant viruses. Commonwealth Mycology Institute/Association of Applied Biologists; Kew, England. 1980.
- Edwardson JR, Christie RG. Alfamovirus Genus. Alfalfa mosaic virus species. In: Edwardson JR, Christie RG, editors. Viruses infecting peppers and other solanaceous crops. University of Florida Press; Gainesville, FL, USA. 1997:63-94.
- Trucco VM, Castellanos CO, Vaghi MCG. Alfalfa Mosaic Virus (AMV): Genetic diversity and a new natural host. J Plant Pathol. 2022;104:349-356.
- Hajimorad MR, Francki RIB. Alfalfa mosaic virus isolates from Lucerne in South Australia: Biological variability and antigenic similarity. Ann Appl Biol. 1988;113:45-54.
- Topkaya S. Molecular characterization of Alfalfa mosaic virus isolates in potato from the Tokat province, Turkiye. Mediterranean Agr Scie. 2022;35(2):75-81.
- Mih AM, Hanson J. Identification of viruses infecting forage grasses in Ethiopia. J Cameroon Acad Scie. 2004;4(3):205-210.
- Oreshkovikj KB, Rusevski R, Kuzmanovska B. Molecular detection and identification of Alfalfa Mosaic Virus (AMV) on pepper cultivated in open fields in Macedonia. Genetika. 2017;49(3):1047-57.
- Moradi A, Almasi MA, Jafary H, Mercado-Blanco J. A novel and rapid loop-mediated isothermal amplification assay for the specific detection of Verticillium dahliae. J of Appl Microb. 2014;116:942-954.
[Crossref] [Google Scholar] [PubMed]
- Davino S, Bivona L, Iacono G, Davino M. First report of Tomato torrado virus infecting tomato in Italy. Plant Dis. 2010;94(9):1172.
[Crossref] [Google Scholar] [PubMed]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63.
[Crossref] [Google Scholar] [PubMed]
- Fernandez-Soto P, Gandasegui AJ, Sanchez HA. A Loop Mediated Isothermal Amplification (LAMP) assay for early detection of Schistosoma mansoni in stool samples: A diagnostic approach in a murine model. PLOS Neg Trop Dis. 2014;8(9):e3126.
[Crossref] [Google Scholar] [PubMed]
- Fu S, Qu G, Guo S. Applications of loop-mediated isothermal DNA amplification. Appl Biochem Biotechnol. 2011;163(7):845-50.
[Crossref] [Google Scholar] [PubMed]
- Przewodowska A, Zacharzewska B, Choluj J. A one-step, real-time reverse transcription loop mediated isothermal amplification assay to detect potato virus Y. Am J Potato Res. 2015;92:303-311.
- Shen W, Tuo D, Yan P. Detection of papaya leaf distortion mosaic virus by reverse-transcription loop-mediated isothermal amplification. J Virol Methods. 2014;195:174-179.
[Crossref] [Google Scholar] [PubMed]
- Wei QW, Yu C, Zhang SY. One-step detection of bean pod mottle virusin soybean seeds by the reverse-transcription loop-mediated isothermal amplification. Virol J. 2012;9:187.
[Crossref] [Google Scholar] [PubMed]
- Zong X, Wang W, Wei H. Rapid detection of Prunus necrotic ringspot virus using magnetic nanoparticle-assisted reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2014;208:85-9.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Gu Q, Sun H. One-step reverse transcription loop mediated isothermal amplification assay for sensitive and rapid detection of cucurbit chlorotic yellows virus. J Virol Methods. 2014 195:63-66.
[Crossref] [Google Scholar] [PubMed]
- Cardosa MJ, Wang SM, Sum MSH Tio PH. Antibodies against prM protein distinguish between previous infection with dengue and Japanese encephalitis viruses. BMC Microbiol. 2002;2(9):1-6.
[Crossref] [Google Scholar] [PubMed]
- Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat Protoc. 2006;1(2):581-85.
[Crossref] [Google Scholar] [PubMed]
- Almasi A, Almasi G. Tracking and identification of Alfalfa Mosaic Virus (AMV) by Loop mediated isothermal amplification assay. Crop Biotech. 2017;18:51-63.
- Liu A, Guan G, Du P, Gou H, Liu Z, Liu J, et al. Loop-Mediated Isothermal Amplification (LAMP) method based on two species-specific primer sets for the rapid identification of Chinese Babesia bovis and B. bigemina. Parasitol Int. 2012;61(4):658-63.
[Crossref] [Google Scholar] [PubMed]
- Warghane A, Misra P, Bhose S. Development of a simple and rapid Reverse Transcription-Loop Mediated Isothermal Amplification (RT-LAMP) assay for sensitive detection of Citrus tristeza virus. J Virol Methods. 2017;250:6-10.
[Crossref] [Google Scholar] [PubMed]
- Chen H, Yan Z, Xuefeng W, Changyong Z, Xiuyan Y, Zhongan L. Detection of citrus yellow vein clearing virus by quantitative real-time RT-PCR. Hortic. Plant J. 2016;2:188-192.
- Garnsey SM. Enzyme-Linked Immunosorbent Assay (ELISA) for citrus pathogens. In: Graft-transmissible diseases of citrus. Hand book for detection and diagnosis. Food and Agriculture Organization of the United Nations, Rome, Italy. 1991:193-216.
- Sharma S, Singh B, Nagpal A. Indexing tools for Indian Citrus Ringspot Virus (ICRSV). Open Biol J. 2009;2:27-31.
Citation: Woubit D, Fikerte M, Yesuf E, Alemayehu TN, Alice M, Chris SJ. (2023) Reverse transcriptase Loop-Mediated Isothermal Amplification assays (RT-LAMP) for the Detection Of Alfalfa Mosaic Virus from Forage Crops, Addis Ababa, Ethiopia. J plant pathol Microbial. 14:690.
Copyright: © 2023 Woubit D, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

