Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 0, Issue 0
Prevalence and Molecular Detection of Escherichia Coli Isolated from Raw Cow Milk at Selected Collection Centers in Bushenyi District, Western Uganda: A Potential Risk to Human Health
Theophilus Pius1*, Blessing J.S. Yashim2, Ibrahim Ntulume2 and Paul Ssajjakambwe32Department of Microbiology and Immunology, Kampala International University, Bushenyi, Uganda
3Department of Veterinary Pharmacy, Kampala International University, Kampala, Uganda
Received: 29-Jul-2024, Manuscript No. JBP-24-26611 ; Editor assigned: 31-Jul-2024, Pre QC No. JBP-24-26611 (PQ); Reviewed: 14-Aug-2024, QC No. JBP-24-26611 ; Revised: 21-Aug-2024, Manuscript No. JBP-24-26611 (R); Published: 28-Aug-2024, DOI: 10.35248/2155-9597.24. S27.101
Abstract
Escherichia coli are known as commensal bacteria in the digestive tract of animals. It is an indicator of faecal contamination when detected in milk, and may be a sign of defective hygiene practices along the processing points. This study aimed to determine the prevalence, molecular characterization, and antimicrobial resistance profiles of Escherichia coli isolated from raw cow milk at selected milk collection centers. The overall prevalence of E.coli in raw milk was 26.7%. The prevalence in the three areas was; Nyakabirizi (42%), Ishaka TC (30%), and Bushenyi TC (10%) with a p-value of 0.022, which is statistically significant to p<0.05, and the posthoc test also showed a significant difference with Nyakabirizi. Of the 23 isolates, 91.3% were susceptible to both chloramphenicol and nalidixic acids. Resistance was observed with trimethoprim-sulfamethoxazole (47%), ampicillin (39.1%), and tetracycline (30.4%). Molecular characterization indicated one sample was positive for the H7 flagella gene after PCR and sequencing.
Keywords
Raw milk, Escherichia coli, Potential risk, Cow, Human
Introduction
Milk is a highly nutritious food recommended to several vulnerable groups, such as young children and expectant mothers [1]. In Sub-Saharan Africa, raw milk is consumed directly by a large number of people in rural areas and indirectly by a much larger segment of the population through the consumption of numerous types of milk products [2]. However, it is prone to contamination by both infectious and non-infectious agents, posing risks to human health. Among some of the infectious agents are Campylobacter spps, Listeria monocytogenes, Salmonella spps, and E.coli [3].
Escherichia coli (E. coli) was first identified as a human pathogen in 1975 from a California patient with diarrhea and was linked to food-borne disease in 1982 [4]. Even though E.coli is a known commensal bacterium as part of the intestinal micro-flora of a variety of animals, not all strains are harmless [5]. Since milk is highly nutritious, several bacteria can multiply within it. The presence of food-borne organisms in milk increases the risk of transmission of food-borne pathogens [6].
Microbes gain entry into milk directly from dairy cows, the farm environment (especially water), equipment used for storage of the milk, and during transportation [7]. Coliforms, E.coli inclusive, are commonly referred to as marker organisms [8].
Among the various E.coli strains, pathogenic E.coli belongs to the first six groups: 1. Enteropathogenic E.coli (EPEC) associated with infantile diarrhoea, 2. Enteroinvasive E.coli (EIEC) causing dysentery-like disease, 3. Enterotoxigenic E.coli (ETEC) produce enterotoxins leading to cases of diarrhea, 4. Enteroaggregative E.coli (EAEC) express aggregative adherence, 5. Diffusely adherent E.coli (DAEC) that adheres to the surface of epithelial cells, and 6. Shiga-producing E.coli (STEC) which may also be referred to as Verocytotoxin-producing E.coli (VTEC) or enterohemorrhagic E.coli (EHEC) produces stx1 and stx 2 causing Hemorrhagic Colitis (HC) and sometimes Hemolytic Uremic Syndrome (HUS) in humans [9].
Once the hygiene at production, handling, transportation, and points of sale of milk is compromised, the milk sold in raw form poses a great risk to public health [10]. This study focused on investigating the presence of E.coli in milk samples collected at selected milk collection centers in the Bushenyi district. There is a paucity of information about the E.coli isolates from milk, with emphasis on the characterization of possible virulence factors at the molecular level.
Materials and Methods
Study area and design
A cross-sectional study was conducted in the Bushenyi district, where raw milk (10 ml) was collected from each center, making a total of 86 collection centers, stratified into three geographical areas (Ishaka town, Nyakabirizi, and Bushenyi trading center), South-western Uganda. This entailed purposive sampling at those centers that consented to participate in this study.
Ethical consideration
Informed consent was obtained after a clear explanation of the study objectives and the benefits. The consent form was administered in either English or Runyankole, the most common indigenous language of the area.
Sample collection and handling
Milk was aseptically collected and placed in sterile sample collection tubes, tightened with a screw cap, and placed on ice in a cool box. The collected samples were transported to the Kampala International University microbiology laboratory for further analysis. Each sample was assigned a serial number immediately after collection, which also concealed the identity of the collection centres to respect the confidentiality agreement in the consent form.
Microbiology culture and isolation
Standard microbiology procedures were employed to run the individual samples. Inoculated plates using the streaking method on VRBA, and MacConkey agar were incubated at 37°C for a maximum period of 48 hours. Bacterial colonies were identified by their common phenotypic characteristics culturally. In addition, conventional biochemical methods were applied to the isolates for further identification of the E.coli isolates.
Briefly, all sample manipulations and preparations were done under a Class 2 biosafety cabinet (Esco, Singapore). A modification of the method used, samples were first enriched in buffered peptone water broth (Oxoid, UK) at 37°C for 24 hours [11]. The overnight bacterial broths were then streaked onto MacConkey Agar (Conda, Spain) and incubated at 37°C for 24 hours. Presumptive E.coli colonies on MacConkey Agar were medium-sized, intense pink (red) in color, and surrounded by a zone of precipitated bile acids. A single colony was further gramstained and sub-cultured on Nutrient Agar to obtain a pure culture before biochemical testing. Biochemical analysis of the E.coli isolates was done using indole, methyl red, lactose and citrate utilization tests.
Sensitivity analysis
Employing the disc diffusion method with modifications, the inocula of isolated strains was prepared in 1% peptone water. Adjustments of the turbidity equal to 0.5 McFarland standard were executed, thereafter, a sample was applied onto Mueller Hinton agar using a sterile swab to spread the culture solution onto the surface of solid media. The inoculated plate was left to dry in a biosafety cabinet for 5 minutes. Subsequently, antibiotic sensitivity discs were applied onto the inoculated agar using sterile forceps and placed in a 37°C incubator. Zones of inhibition were measured in mm with the help of a ruler after 18-24 hours of incubation and findings were compared against a reference chart. The antibiotics used were ampicillin (30 μg), tetracycline (30 μg), chloramphenicol (30 μg), neomycin (30 μg), nalidixic acid (30 μg) and trimethoprim-sulfamethoxazole (5 μg) [12].
Polymerisation chain reaction
A conventional Polymerase Chain Reaction (PCR) test was run for the E.coli isolates to check for the presence of the gene specifically coding for the flagella antigen H7. The primer sequences were Forward-GCGCTGTCGAGTTCTATCGAGC and Reverse-AACGGTGACTTTATCGCCATTCC, adopted and the expected band size was 625 base pairs [13]. For a 25 μl reaction volume, 15 μl of a one Taq PCR master mix was used as per manufacturer’s instructions, with 1 μl of 10 μM of each primer, 1 μl DNA template and 7 μl of PCR grade water [2,15]. The initial denaturation temperature was 95°C for 5 minutes, followed by denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, and 72°C for 1 minute, all run for 30 cycles. The final extension was at 72°C for 10 minutes and a holding temperature of 4°C. The PCR products were loaded on an agarose gel and viewed under UV light after electrophoresis [14].
Results
Prevalence
In this study, selected collection centers indicated the presence of E.coli in the milk (a threat to human health), with a p-value of 0.02, which is a statistically significant finding (Table 1).
| Study areas | No. of Samples | Prevalence (%) | 95% CI | P value | |
|---|---|---|---|---|---|
| LL | UP | ||||
| Ishaka TC | 30 | 9 (30) | 14.73 | 49.40 | 0.02 |
| Nyakabirizi | 26 | 11 (42.3) | 23.37 | 63.08 | |
| Bushenyi TC | 30 | 3 (10) | 2.11 | 26.53 | |
| Total | 86 | 23 (26.74) | 17.77 | 37.38 | |
Table 1: Presence of E.coli in the milk in selected collection centers.
Colony-Forming Units (CFUs)
The Colony Forming Unit (cfu) enumeration revealed the total bacteria counts of the different study areas with Bushenyi and Nyakabiizi districts having the highest counts above the internationally acceptable standard of 3.62 × 107 ± 1.37 × 107 cfu/ml (Table 2).
| Sampling areas | Total aerobic counts (cfu/ml) | One sample t-test | ||||
|---|---|---|---|---|---|---|
| Minimum | Maximum | Mean ± SD | 95% CI | t-value | P-value | |
| CFUs (x107) | ||||||
| Ishaka TC | 0.026 | 19.3 | 2.39 ± 3.61a | 1.037-3.735 | 3.617 | 0.001 |
| Nyakabirizi | 0.029 | 17.3 | 6.15 ± 5.48b | 3.929-8.357 | 5.716 | <0.0001 |
| Bushenyi TC | 0.011 | 58.0 | 6.17 ± 1.29b | 1.362-10.981 | 2.624 | 0.014 |
Table 2: Multiple Tukey comparison test with significant differences (p<0.05) represented by different superscripts (a, b). TCTrading center.
Antimicrobial resistance profiles
Among the isolates, ampicillin and tetracycline showed the highest resistance as seen (Table 3 and Figure1).
| Number of E.coli isolates (n=23) | |||
|---|---|---|---|
| Antibiotic used | Susceptible | Intermediate | Resistant |
| Ampicillin (30 µg) | 7 (30.4%) | 7 (30.4%) | 9 (39.1%) |
| Tetracycline (30 µg) | 9 (39.1%) | 7 (30.4%) | 7(30.4%) |
| Neomycin (30 µg) | 0 (0%) | 18 (78.3%) | 5 (21.7%) |
| Chloramphenicol (30 µg) | 21 (91.3%) | 0 (0%) | 2 (8.7%) |
| Nalidixic acid (30 µg ) | 21 (91.3%) | 2 (8.7%) | 0 (0%) |
| Trimethoprim-sulfamethoxazole (5 µg) | 12 (52.2%) | 0 (0%) | 11 (47.8%) |
Table 3: Antibiotic used for E.coli isolates.
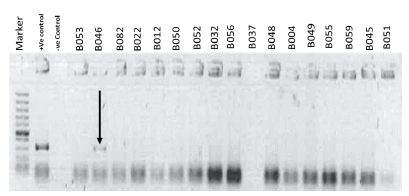
Figure 1: A conventional PCR on the E.coli isolates showed one of the isolates as positive for the gene coding for the flagella antigen H7 (B046), having the required band of 625 bps indicated by the black arrow.
Discussion
Milk is an essential supporting medium for microbial growth. Food-borne infections are caused by a wide range of microbial agents, such as Campylobacter, Salmonella, Escherichia, Brucella, and Staphylococcus, among others which result in mild or lifethreatening illnesses. The main objective of this study was to carry out a baseline survey to assess the possible extent of E.coli contamination of cow milk at selected collection centers in the Bushenyi district. This was against the backdrop that consumption of unpasteurized milk either directly or indirectly had been reported to be a threat to human health [15].
In this study, E.coli was isolated from raw milk, (Table 1). Such a finding is suggestive of fecal contamination, from either water, the environment, or the animals. Another study suggested that contamination of milk and its products had a bearing on the housing conditions, and hygiene standards at milk handling, transportation, and storage [16]. This implied a potential for food-borne illness among consumers of the affected raw milk was eminent. Similar studies revealed prevalence ranging from 7%-90% [17-19].
The CFU counts of Bushenyi and Nyakabirizi districts (Table 2) were above the acceptable cutoff (3.62 x 107 ± 1.37 x 107 cfu/ ml.). For milk, thus a further indication of a threat to human health just in case such milk is not cooked well to kill the bacteria present before consumption. Oltramari et al., and Ribeiro et al., revealed high bacteria counts, this attributable to poor hygiene [2,20]. Furthermore, a pilot study before this work observed that the absence of a milking paler, and scarcity of clean water among others could also contribute to the above findings. In addition, it was noted that the injudicious use of antibiotics by farmers to treat a range of conditions even without appropriate professional guidance from a veterinarian was a common practice. The latter was raised thus a fertile ground to contribute to the development of antibiotic resistance [21]. The trends in (Table 3) were no surprise, with the major resistance noted against ampicillin and tetracycline. Tetracycline is one of the most abused drugs by livestock communities, whereby it is used to treat a range of infections. Since the responses are quite commendable, it is a common drill to carry this drug in the first aid kit of many farmers. The use of tetracycline for prophylactic purposes by these communities is the order of the day. A study showed a resistance of 100% of E.coli to tetracycline [22]. These two drugs are very common antibiotics that farmers use indiscriminately in the forms of penstrep and oxytetracycline to treat almost any thought of infection in animals. That said, the likely development of resistance by the prevalent bacteria may not be uncommon.
Furthermore, molecular characterization to investigate the presence of the gene in the E.coli isolates that codes for the flagella antigen H7 was performed. Only one sample (B046) was positive. Also to note, earlier and similar work by Msolo et al., and Lye et al., identified this gene that codes for the organelle, the flagella used for motility and adherence onto host cells [5,19]. It’s known that E.coli strains that possess an active gene coding for the flagella antigen H7 are more virulent than those that do not express this gene. This further implies that once humans consume milk from the B046 collection center, chances of developing food-borne infections are high once the individuals' milk is not well prepared, and also if all the host immune system is compromised, such as in HIV/AIDS patients.
Conclusion
The presence of E.coli in milk, especially in amounts above the accepted, and also if the isolates do possess known virulence factors as a flagellum, poses a risk to human health. This calls for appropriate preparation of food before consumption in addition to adherence to the required hygiene standards of handling and processing of food destined for human consumption. Additionally, the use of antibiotics after the guidance of a trained medical practitioner is highly recommended.
References
- Lucey JA. Raw milk consumption: Risks and benefits. Nutr today. 2015;50(4):189-93.
- Oltramari K, Cardoso RF, Patussi EV, Santos AC, Mikcha JM. Genetic heterogeneity of Escherichia coli isolated from pasteurized milk in State of Paraná, Brazil. B J Pharma Sci. 2014;50(2):337-43.
- Knight JTJ, Hang’ombe MB, Songe MM, Sinkala Y, Grace D. Microbial contamination and hygiene of fresh cow’s milk produced by smallholders in Western Zambia. Int J Environ Res Public Health. 2016;13(7):737.
- Doyle ME, Archer J, Kaspar CW, Weiss R. Human illness caused by E.coli O157: H7 from food and non-food sources. FRI Briefings. 2006;10(8):2013.
- Msolo L, Igbinosa EO, Okoh AI. Prevalence and antibiogram profiles of Escherichia coli O157: H7 isolates recovered from three selected dairy farms in the Eastern Cape Province, South Africa. Asian Pacific J Trop Dis. 2016; 6(12):990-5.
- Spano LC, Da CKF, Monfardini MV, Cássia BFR, Scaletsky IC. High prevalence of diarrheagenic Escherichia coli carrying toxin-encoding genes isolated from children and adults in southeastern Brazil. BMC Infect Diseases. 2017; 17:1-9.
[Crossref] [Pubmed][Google Scholar]
- Fadaei A. Bacteriological quality of raw cow milk in Shahrekord, Iran. Veterinary World. 2014;7(4).
- Sudda MM, Mtenga AB, Kusiluka LJ, Kassim N. Prevalence and antibiotic susceptibility of Escherichia coli and Salmonella spp. isolated from milk of zero grazed cows in Arusha City. Afr J Microbiol Res. 2016;10(46):1944-51.
- Tamura K, Sakazaki R, Murase M, Kosako Y. Serotyping and categorisation of Escherichia coli strains isolated between 1958 and 1992 from diarrhoeal diseases in Asia. J Med Microbiol . 1996;45(5):353-8.
- Shahzad KA, Muhammad K, Sheikh AA, Yaqub T, Rabbani M, Hussain T, et al. Isolation and molecular characterization of shiga toxin producing E coli O157. 23(6):1618–21.
- Blanco CX, Bonino MP, Sanin MS, Petrina JF, Disalvo VN, Massa R, et al. Potential zoonotic pathovars of diarrheagenic Escherichia coli detected in lambs for human consumption from Tierra del Fuego, Argentina. Microorganisms. 2021;9(8):1710.
[Crossref] [Pubmed][Google Scholar]
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am Clinical Path. 1966;45(4):493-6.
[Crossref] [Pubmed][Google Scholar]
- Chase TME, Rosser T, Allison LJ, Courcier E, Evans J, McKendrick IJ, et al. Pathogenic potential to humans of bovine Escherichia coli O26, Scotland. Emerg Inf Dis. 2012;18(3):439.
[Crossref] [Pubmed][Google Scholar]
- Islam MA, Mondol AS, De Boer E, Beumer RR, Zwietering MH, Talukder KA, et al. Prevalence and genetic characterization of shiga toxin-producing Escherichia coli isolates from slaughtered animals in Bangladesh. Appl Environ Microbiology. 2008;74(17):5414-21.
[Crossref] [Pubmed][Google Scholar]
- Neher S, Hazarika AK, Sharma RK, Barkalita LM, Bora M, Deka P. Detection of Shiga-toxigenic Escherichia coli in milk samples of cattle by PCR. J Agric Veter Sci. 2015;8(5):75-8.
- Reta MA, Addis AH. Microbiological quality assessment of raw and pasteurized milk. Int J Food Sci Microbiology. 2015;2(6):087-91.
- Kateete DP, Kabugo U, Baluku H, Nyakarahuka L, Kyobe S, Okee M, et al. Prevalence and antimicrobial susceptibility patterns of bacteria from milkmen and cows with clinical mastitis in and around Kampala, Uganda. PloS one. 2013;8(5):e63413.
[Crossref] [Pubmed][Google Scholar]
- Lubote R, Shahada F, Matemu A. Prevalence of Salmonella spp. and Escherichia coli in raw milk value chain in Arusha, Tanzania. 2014.
- Lye YL, Afsah HL, Chang WS, Loo YY, Puspanadan S, Kuan CH et al. Risk of Escherichia coli O157: H7 transmission linked to the consumption of raw milk. Int Food Res J. 2013;20(2):1001.
- Ribeiro LF, Barbosa M, Pinto FR, Lavezzo LF, Rossi GA, Almeida H, et al. Diarrheagenic Escherichia coli in raw milk, water, and cattle feces in non-technified dairy farms. Ciência Animal Brasileira. 2019;20.
- Ssajjakambwe P, Bahizi G, Setumba C, Kisaka SM, Vudriko P, Atuheire C, et al. Milk hygiene in rural southwestern Uganda: Prevalence of mastitis and antimicrobial resistance profiles of bacterial contaminants of milk and milk products. Vet Med Int. 2017;2017(1):8710758.
[Crossref] [Pubmed] [Google Scholar]
- Khairy RM, Fathy ZA, Mahrous DM, Mohamed ES, Abdelrahim SS. Prevalence, phylogeny, and antimicrobial resistance of Escherichia coli pathotypes isolated from children less than 5 years old with community acquired-diarrhea in Upper Egypt. BMC infectious diseases. 2020 ;20:1-9. 1.
[Crossref] [Pubmed] [Google Scholar]
Citation: Pius T, Yashim BJS, Ntulume I, Ssajjakambwe P (2024). Prevalence and Molecular detection of Escherichia coli isolated from raw cow milk at selected collection centres in Bushenyi district, Western Uganda: A potential risk to human health. J Bacteriol Parasitol. S27:101.
Copyright: © 2024 Pius T. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.