Indexed In
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 3, Issue 1
Phenotypic and Molecular Study of Carbapenemase-Producing Enterobacteriaceae in a Regional Hospital in Northern Morocco
Younes Mahrach1,2, Nadira Mourabit1,2, Abdelhay Arakrak1, Mohammed Bakkali1 and Amin Laglaoui1*2Mohamed V Hospital, Microbiology Laboratory, Tangier, Morocco
Received: 23-Apr-2019 Published: 17-May-0209
Abstract
Objective: This study aims to assess the prevalence of clinical isolates of carbapenemase-producing Enterobacteriaceae (CPE), and to characterize the types of carbapenemases produced among the strains isolated from different samples in patients hospitalized at the regional hospital in Tangier in the North of Morocco over 12 month period (January to December 2015).
Methods: A total of 367 isolates of Enterobacteriaceae were collected from inpatients, antimicrobial susceptibility of the isolates was determined, and Enterobacteriaceae isolates with reduced susceptibility to carbapenems were investigated by phenotypic study using a range of techniques including antibiotic susceptibility testing, modified Hodge test and the double-disk synergy method. Expression of the genes encoding carbapenemases measured using PCR.
Results: Twenty-two strains of Enterobacteriaceae were isolated which were expressing the phenotypic production of carbapenemases so 5.99% of Enterobacteriaceae are CPE. According to molecular study, the blaOXA-48 gene was the most commonly encountered with twelve isolates harboring this gene. Two isolates were carrying the blaIMP-1 gene, two carried the blaVIM-1 gene, a further two carried the blaKPC-1 gene, and two strains were found to be harboring more than one gene.
Conclusion: This is the first study to reveal the prevalence of CPE in Northern Morocco and reports the first description of Metallo-beta-lactamase KPC- 1-producing Enterobacteriaceae in Morocco.
Keywords
Carbapenemases; Enterobacteriaceae; blaOXA-48; blaIMP-1; blaVIM-1; blaKPC-1
Introduction
The interest shown in recent years in bacterial infections caused by multidrug-resistant bacteria (MDRB), especially carbapenemase-producing Enterobacteriaceae (CPE), by the pharmaceutical companies in charge of therapeutics is still relevant.
Indeed, these infections have clinical importance as they constitute a major public health risk due to the difficulty of their treatment.
Many authors have been interested in evaluating the infections of bacteria producing carbapenemases in various aspects in the epidemiological, clinical or even therapeutic occurrence in order to fight against them.
To our knowledge, few studies report the epidemiological and molecular aspects of these infections in Morocco.
Our work aims to establish the prevalence of and to study the strains of carbapenemase-producing Enterobacteriaceae (CPE) found in patients in the Mohamed V regional hospital in Tangier in the north of Morocco.
Materials and Methods
Study framework
This was a 12-month prospective study, carried out between January and December 2015, of 2178 bacteriological examinations carried out in the microbiology laboratory of the Mohamed V regional hospital in Tangier. The bacterial strains were isolated from urine, blood, pus, bone samples, sputum, pleural fluid, and peritoneal fluid, obtained from in patients hospitalized at the regional hospital in Tangier in the North of Morocco.
Enterobacteriaceae identification
The bacterial culture was grown on cysteine lactose electrolyte deficient (CLED) agar and eosin methylene blue (EMB) media, and the identification of the Enterobacteriaceae was carried out using the usual techniques such as Gram staining, oxidase and Api 20E gallery (BioMérieux-France) following the recommendations of the Clinical and Laboratory Standards Institute CLSI [1].
Antibiotics tested
The antibiotic sensitivity study was carried out by the Kirby- Bauer method on Muller-Hinton agar medium according to the CLSI [1] guidelines. The eighteen antibiotics tested were: Amoxicillin/clavulanic acid (25/10 30 μg), Ceftaxidine (30 μg), Ceftazidime (30 μg), Cefixime (10 μg), Ceftriaxone (30 μg), Colistin (50 μg), Cefotaxime (10 μg), 10 mg), Norfloxacin (10 μg), Trimethoprim/Sulfamethoxazole (1 μg), Fosfomycin (50 μg), Gentamycin (10 μg), Piperacillin/ tazobactam (25/23.75 μg), Trimethoprim (5 μg). A Cloxacillin disk (5 μg) was also used for hyperproduction of cephalosporinases with porin loss [1,2]. The E. coli strain ATCC 25922 was used as an internal quality control.
Phenotypic characterization of carbapenemase-producing enterobacteria
The detection of carbapenemase-producing bacteria was carried out using the Kirby-Bauer disk diffusion method using 10 μg ertapenem discs, 10 μg meperenem and 10 μg imipenem, on Muller-Hinton agar. Any strain having a decreased sensitivity to Ertapenem (inhibition zone <22 mm) was considered suspicious of being carbapenemase producing and was verified by other phenotypic tests as recommended by CLSI [1].
Modified Hodge Test (MHT): A 0.5 MacFarland turbidity suspension of the E. coli strain E. coli ATCC 25922 was prepared and inoculated onto Muller Hinton Agar medium. The medium was dried and a disk of ertapenem (10 μg) or meropenem (10 μg) was placed in the center of the test zone. Using a straightline swab, the test strain was distributed from the middle edge to the center and this was repeated with each of the test strains plus the control strains (positive and negative) in different directions. The samples were incubated overnight at 35°C +/- 2°C. The results were read after incubation; if bacterial growth in the form of flooding occured at the intersection of the E. coli 25922 inhibition zone with the line of the test strain then the test was considered positive, otherwise the test was considered negative for the production of carbapenemases [3].
Phenotypic inhibition tests: In short, they are based on the inhibitory properties of boronic acid or clavulanic acid with respect to carbapenemases of class A, and cloxacillin with respect to cephalosporinases. Using a cloxacillin disk helps to differentiate the strains which have carbapenem sensitivity from those which exhibit resistance by production of low-level carbapenemases [2,4], while the detection of metallo-betalactamase production was carried out by the disk test combined with two imipenem discs (10 μg), one of which contained 10 μg of anhydrous 0.1 M (292 μg) EDTA (Sigma Chemicals, St. Louis, MO). The disks were placed at an interval of 25 mm on a Muller-Hinton agar box, an increase in the inhibition diameter of the zone of > 4 mm around the imipenem-EDTA disk compared to that of the imipenem disc alone was considered positive for the production of metallo-beta-lactamase [5].
Detection of ESBL: The detection of extended spectrum betalactamases (ESBL) is based on the study of the synergy between the disks of amoxicillin + clavulanic acid (AMC) and third generation C3G cephalosporins, ceftazidime, a cefepime C4G disk and a Monobactam aztreonam disk (Figure 1) [6].

Figure 1: Photographic image of an electrophoresis gel of amplicons of the blaOXA-48 gene, the size of which is 744 kb, this image shows from left to right six isolates (17, 18, 19, 20, 21 and 22) carrying the BlaOXA-48 gene TN: negative control and PM: weight markers.
Search for coding genes for carbapenemases
Extraction of DNA: Extraction of the chromosomal DNA was done by boiling from a bacterial culture in the stationary phase (16-24 h) on a solid medium at 37°C. Two colonies were suspended in 100 μl of water distilled RNase and DNase free waterthen vortexed for 10 seconds. The preparation was placed in a water bath at 100°C for 10 minutes and then stored at -20°C until use. The samples were centrifuged at 19000xg for 5 min. 2 μl of extracted DNA was used as a PCR matrix [7], while extraction of the plasmid DNA was done using a commercial kit (Bioline Isolate plasmid Mini Kit Cat No. Bio-52026) according to the manufacturer's instructions.
Amplification of target genes: The polymerase chain reaction (PCR) technique was used to amplify the genes encoding (class-1 blaNMC-1, blaKPC-1) or serine-carbapenemases according to Ambler's classification (blaIMP-1, blaVIM-1) Class B or metallocarbapenemase (blaOXA-48) class D or oxacillinases and the integron gene. The primers used for PCR are listed in Table 1. PCR amplifications were performed in 50 μL final volumes with 1 x reaction buffer, 0.2-0.4 U Taq polymerase, 200 μM of each dNTP, 1.5 mM MgCl2 and 200 nM of each primer. 2 μL of DNA was extracted using the boiling technique or 1 μL prepared per kit.
| Type of enzyme | Amorces | Séquence des amorces | Taille (pb) | Support | Reference |
|---|---|---|---|---|---|
| KPC-1 | KPC-1 F | ATGTCACTGTATCGCCGTCT | 893 | Plasmidique | [8] |
| KPC-1 R | TTTTCAGAGCCTTACTGCCC | ||||
| NMC-1 | NMC-1 F | GCATTGATATACCTTTAGCAGAGA | 2178 | chromosomique | [8] |
| NMC-1 R | CGGTGATAAAATCACACTGAGCATA | ||||
| IMP-1 | IMP-1 F | TCGTTTGAAGAAGTTAACGG | 568 | Plasmidique | [13] |
| IMP-1 R | ATGTAAGTTTCAAGAGTGATGC | ||||
| VIM-1 | VIM-1 F | GGTGTTTGGTCGCATATCGCAA | 520 | Plasmidique ou chromosomique |
[13] |
| VIM-1 R | ATTCAGCCAGATCGGCATCGGC | ||||
| OXA-48 | OXA-48 F | TTGGTGGCATCGATTATCGG | 744 | Plasmidique | [8] |
| OXA-48R | GAGCACTTCTTTTGTGATGGC | ||||
| Intégrons 5' | GGCATCCAAGCAGCAAG | variable | Plasmidique ou chromosomique |
[8] | |
| Intégrons 3' | AAGCAGACTTGACCTGA |
Table 1: Oligonucleotides used for gene amplification in this study.
The conditions of the PCR were as follows: initial denaturation at 94°C for 2 min then denaturation at 94°C for 30 s, hybridization of the two primers taking place at a variable temperature according to the primers for 45 s, extension at 72°C for 1 min; and the final extension at 72°C for 7 min, for 35 cycles.
The products of the amplification of the target genes were revealed by electrophoresis on a 1% agarose gel supplemented with ethidium bromide.
Results
Out of the 2178 samples, 503 (23.1%) samples met the 105 UFC [1] criteria, with 367 (72.96%) Enterobacteriaceae. The most identified bacteria among these Enterobacteriaceae were:
E. coli 264, ie 71.9% of Enterobacteriaceae, Klebseilla pneumoniae 84 or 22.8%.
The results of the antibiogram identified 22 strains (5.99%) which were resistant to at least ertapenem and validating the phenotypic tests which indicate a probable production of carbapenemases.
Resistance to carbapenems was associated with resistance to other families of antibiotics; among these twenty-two strains, seven were identified as producing both ESBL and carbapenemases (Table 2).
| Bacteria | Origin | The Antibiotic Test | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CN | FF | NOR | CT | ETP | IPM | MEM | TPZ | ATM | AMC | FOX | CFM | CTX | CAZ | CRO | FEP | TIM | SXT | NB | ||
| Klebsiella pneumonie | Pus | S | R | S | R | R | S | S | S | S | R | S | R | S | S | S | S | R | R | |
| Proteus mirabilis | CBU | S | S | R | R | R | S | S | S | S | R | S | R | S | S | S | S | R | S | |
| E. coli | CBU | S | R | R | R | R | S | S | S | S | R | R | R | S | S | S | S | R | R | |
| E. coli | CBU | S | S | R | R | R | S | S | S | R | R | R | R | R | S | R | R | R | R | |
| E. coli | Pus | S | S | S | R | R | S | S | S | S | R | S | S | S | S | S | S | R | R | |
| E. coli | Pus | S | S | S | R | R | S | S | S | S | R | R | R | S | S | S | S | R | R | |
| Providencia | Pus | R | S | R | R | R | S | S | S | R | R | R | R | S | S | S | R | R | S | |
| Enterobacter colaceae | CBU | S | S | R | R | R | S | S | S | S | R | R | R | R | R | R | S | R | S | BLSE |
| E. coli | CBU | S | R | R | R | R | S | S | S | R | R | R | R | R | R | R | R | R | R | |
| E. coli | Pus post op | R | S | R | R | R | S | S | S | S | R | R | R | S | S | S | S | R | S | |
| Enterobacter Cloaeae | Pus post op | R | S | R | R | R | S | S | R | R | R | R | R | R | R | R | R | R | R | |
| Enterobacter colaceae | Pus post op | R | R | R | R | R | R | S | R | R | R | R | R | R | R | R | R | R | R | |
| E. coli | Pus post op | R | R | R | R | R | R | S | R | R | R | S | R | R | R | R | R | R | S | |
| Klebsiella pneumonie | Pus post op | R | S | R | R | R | S | S | S | R | R | R | R | R | R | R | R | R | R | |
| Klebsiella pneumonie | Pus | R | S | R | R | R | S | S | R | R | R | R | R | R | R | R | R | R | R | BLSE |
| E. coli | Pus post op | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R | |
| Enterobacter colaceae | Pus post op | R | S | R | S | R | S | S | S | S | R | S | R | S | S | S | S | R | R | |
| Klebsiella pneumonie | Pus post op | R | S | R | S | R | S | S | S | R | R | R | R | R | R | R | R | R | R | BLSE |
| Klebsiella pneumonie | Pus post op | R | S | R | S | R | S | S | S | S | R | R | R | R | R | R | R | R | R | BLSE |
| Klebsiella pneumonie | Pus post op | S | S | R | S | R | S | S | S | R | R | R | R | R | R | S | S | R | R | BLSE |
| E. coli | CBU | R | S | R | S | R | S | S | S | R | R | R | R | R | R | R | R | R | R | BLSE |
| Klebsiella pneumonie | CBU | R | S | R | R | R | S | S | R | R | R | R | R | R | R | R | R | R | R | BLSE |
AMC: Amoxicillin/clavulanic acid (25/10 μg), ATM: Aztreonam (30 μg), CAZ: Ceftazidime (30 μg), CFM: Cefixime (10 μg), CRO: Ceftriaxone Μg), CTX: Cefotaxime (30 μg), FEP: Cefepime (30 μg), FOX: Cefoxitin (30 μg), ETP: Ertapenem (10 μg), FF: Fosfomycin TIM: Trimethoprim (5 μg), ESB: Piperacillin/tazobactam (75/10 μg), IPM: Imipenem, MEM: Meropenem, NOR: Norfloxacin, SXT: Trimethoprim/Sulfamethoxazole, ELBS beta-lactamases with broad spectrum.
Table 2: Resistance profile of suspected EPC strains to antibiotics recommended by CLSI.
After amplification of the target genes, there were 12 cases of the blaOXA48 gene (Figure 2), two cases of the bla-KPC-1 gene, two cases of the bla-VIM-1 gene, and two cases of the blaIMP-1 gene.
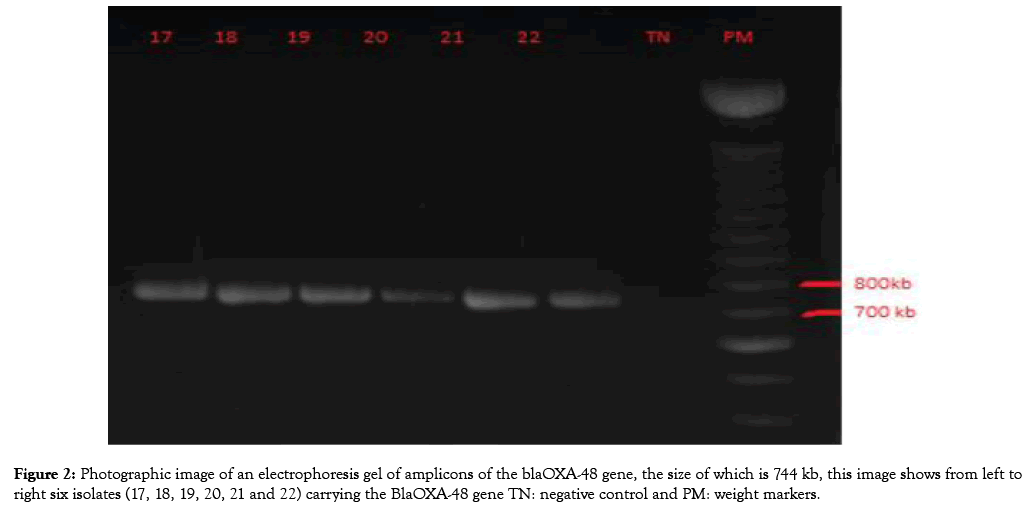
Figure 2: Image showing the synergy between the AMC disk at the center and the C3G discs (CTX and CAZ), the C4G (FEP) disc and the aztreonam (ATM).
There was also one case which contained both the blaOXA-48 and blaKPC-1 genes, and one case which harbored the integron gene plus the blaOXA-48 gene. A further two cases of the integrons gene have been recorded while none of the strains carrying the blaNMC-1 genes, and the remainder of the strains were negative for the subject genes (Table 3).
| S.NO. | Bacteria | KPC | GES | NMC | VIM | IMP | OXA-48 | Integron | Remarks |
|---|---|---|---|---|---|---|---|---|---|
| 1 | K. pneumoniae | + | + | ||||||
| 2 | P. mirabilis | ||||||||
| 3 | E. coli | + | |||||||
| 4 | E. coli | + | |||||||
| 5 | E. coli | + | |||||||
| 6 | E. coli | + | |||||||
| 7 | Providencia | ||||||||
| 8 | Enterobacter C | + | BLSE | ||||||
| 9 | E. coli | + | |||||||
| 10 | E. coli | ||||||||
| 11 | Enterobacter C | + | |||||||
| 12 | Enterobacter C | + | + | BLSE | |||||
| 13 | E. coli | + | |||||||
| 14 | K. pneumoniae | + | |||||||
| 15 | K. pneumoniae | + | BLSE | ||||||
| 16 | E. coli | BLSE | |||||||
| 17 | Enterobacter C | BLSE | |||||||
| 18 | K. pneumoniae | + | BLSE | ||||||
| 19 | K. pneumoniae | + | BLSE | ||||||
| 20 | K. pneumoniae | + | |||||||
| 21 | E. coli | + | |||||||
| 22 | K. pneumoniae | + | + | ||||||
| TOTAL | 22 | 2 | 0 | 0 | 2 | 2 | 14 | 2 | 7 |
Table 3: Distribution of the genes encoding the carbapenemases recorded according to the strains.
Discussion
The enormous use of carbapenems in the prevention and treatment of infections caused by multiresistant bacteria, especially Enterobacteriaceae producing extended-spectrum betalactamases (ESBL) [8], causes the emergence of carbapenemresistant Enterobacteriaceae. Resistance to carbapenems may involve several combined mechanisms but the production of enzymes remains the most worrying mechanism because of the genetic supports that can be transferred horizontally as well as vertically. Since the first identification of carbapenemases (NMC) in Enterobacteriaceae in 1993 [9], carbapenemases have been widely reported in Enterobacteriaceae worldwide [9]. In Morocco the trend is similar [9] as several studies conducted in regions in central Morocco have addressed the problem of the appearance of CPE in both hospitals and the community [10] and even the environment [11].
However, no studies of CPE have been conducted in the northern region. Our study is the first survey of carbapenemresistant Enterobacteriaceae, especially CPEs in the Tangier region of northern Morocco, and provides information on the magnitude of the problem from an epidemiological and microbiological point of view in this area.
In this study, we identified 367 (72.96%) Enterobacteriaceae of which 22 were hospital-resistant strains (5.99%) which were resistant to carbapenems and validating phenotypic tests indicated a probable production of carbapenemase. Molecular tests confirmed the production of carbapenemases in sixteen cases; twelve OXA-48, two KPC-1, two VIM-1, and two IMP-1. There were two strains, K. pneumoniae and Enterobacter Colaceae, which carried two genes at once bla OXA-48 and blaKPC-1.
The results of the molecular tests are consistent with those of the phenotypic tests for showing that they are the enterobacteria producing the carbapenemases with a broad diffusion of the enzyme of class D beta-lactamase OXA-48. Here we report 12 isolates producing this enzyme which accounts for 75% of the CPE detected since the first OXA-48 enzyme description in 2001 in Turkey [9]. This type of carbapenemase has spread widely throughout the world [10]. In Morocco the first case was reported in recent years, and the emergence and spread of the OXA-48 enzyme has been widely reported in several Moroccan hospitals. The enzyme OXA-48 has been a source of nosocomial infection in many parts of the world. Recently, OXA-48 has been identified in a number of Mediterranean countries as an endemic; such is the case for Morocco, Tunisia and Turkey [12,13].
We also note the presence of two class A KPC-1 CPEs and to our knowledge no cases have been recorded in Morocco so far; those are the K. pneumoniae and an Enterbacter cloaceae strains from two different patients who were hospitalized after surgery in the traumatology department.
Two cases of IMP-1 metallo-beta-lactamase enzymes have been reported in an isolate of E.coli and isolate of K. pneumoniae and two cases of VIM-1 metallo-beta-lactamase enzymes were detected by PCR in all 22 phenotypically positive isolates. The latter was described for the first time in Morocco in a strain of Klebseilla by Barguiga et al. in 2012 [14]. In our case it was recorded in a strain of E. coli and one Enterobacter cloaceae from two hospitalized patients.
This study shows a prevalence of 5.99% of the producer carbapenemase strains. This prevalence is high compared to the prevalence of 2.8% described by Wartiti M.Al et al. in another region of Morocco [15,16].
The 22 strains isolated are of hospital origin, they are all isolated from the surgical services, with the patients in postoperative which is explained by the nosocomial origin of the infection [17].
Seven carbapenemase producing strains are associated with ESBL. Resistance of bacteria to carbapenem in association with resistance to other families of antibiotics, especially in cases of ESBL, could lead to therapeutic impasses [18].
Conclusion
The increasing occurrence of producing carbapenemase strains represents a public health risk that deserves proper monitoring or even the implementation of a national program to combat BMR in order to avoid the emergence of an unbeatable bacterial generation.
Future Work
This study will be supplemented by other molecular techniques in order to continue the specification of the genes responsible to produce carbapenemases. A time extension of this study is also planned.
Conflict of Interest
All authors declare to have no conflict of interest
Ethical Approval
Not required.
REFERENCES
- Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement (2001) CLSI document M100-S24. Wayne, PA: CLSI.
- Giske CG, Gezelius L, Samuelsen Ø, Warner M, Sundsfjord A, Woodford N. A sensitive and specific phenotypic assay for detection of metallo‐β‐lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect. 2017;17(4):552-556.
- Clare F, Lisa L, Anton YP. Phenotypic detection of carbapenem susceptible metallo-β-lactamase-producing gram-negative bacilli in the clinical laboratory. J Clin Microbiol. 2006;44(9):3139-3144.
- Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V. Identification and screening of carbapenemase‐producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18(5):432-438.
- Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobactet species. Clin Microbiol Infect 2001;7(2):88-91.
- Garrec H, Drieux RL, Golmard JL, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of extended-spectrum β-lactamase production by Enterobacteriaceae. J Clin Microbiol. 2011;49(3):1048-1057.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual (No. Ed. 2). Cold spring harbor laboratory press 1989;58: xxxviii + 1546.
- Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15-22.
- Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. 2007;20(3),440-458.
- Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 2017;67(7):1597-1606.
- Barguigua A, El Otmani F, Talmi M, Zerouali K, Timinouni M. Emergence of carbapenem-resistant Enterobacteriaceae isolates in the Moroccan community. Diagn Microbiol Infect Dis. 2012;73(3):290-291.
- El wartiti MA, Bahmani FZ, Elouennass M, Benouda A. Prevalence of Carbapenemase-Producing Enterobacteriaceae in a University Hospital in Rabat, Morocco: A 19-Months Prospective Study. Int Arab J Antimicrob Agents. 2012;2(3).
- Benouda A, Touzani O, Khairallah MT, Araj GF, Matar GM. First detection of oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Morocco. Ann Trop Med Parasitol. 2010;104(4),327-330.
- Barguigua A, Talmi M, Zerouali K, Timinouni M. First report of a Klebsiella pneumoniae strain coproducing NDM‐1, VIM‐1 and OXA‐48 carbapenemases isolated in Morocco. APMIS 2013;121(7):675-677.
- Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413-431.
- Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae Emerg Infect Dis. 2011;17(10):1791-1798.
- Potron A, Poirel L, Bussy F, Nordmann P. Occurence of the carbapenem-hydrolyzing β-lactamase gene blaOXA-48 in the environment in Morocco. Antimicrob Agents Chemother. 2011;55(11): 5413–5414.
- Zong Z, Lü X, Valenzuela JK, Partridge SR, Iredell J. An outbreak of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in western China. Int J Antimicrob Agents. 2008;31(1):50-54.
Citation: Mahrach Y, Mourabit N, Arakrak A, Bakkali M, Laglaoui A (2019) Phenotypic and Molecular Study of Carbapenemase-Producing Enterobacteriaceae in a Regional Hospital in Northern Morocco. J Clin Med Sci. 3:113. doi: 10.35248/2593-9947.19.3.113
Copyright: © 2019 Mahrach Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
