Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Scimago
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- MIAR
- University Grants Commission
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 0, Issue 0
Peptide-mRNA Vaccine for SARS-Cov-2
Esmaeil Farshi*Received: 03-Sep-2020 Published: 24-Oct-2020, DOI: 10.35248/2157-7560.20.S4.002
Abstract
A vaccine is described which elicits a protective immune response against SARS-Cov-2. Designed engineered peptides that could strongly bind to the spike protein of coronavirus inside cells, and to use these peptides to trigger the cells to break down the viral proteins. In this vaccine the mRNA (encoding Ribonucleic Acid) encodes a stable perfused form (the form before being fused to the cell membrane of the host cell) of Spike protein (S). In this vaccine the main problem of short time remaining of antibodies of SARS-Cov-2 virus in body could be compensated by activation of T cells that causes long term immunity because memory T cells remain in body at least 11 years. A peptide based vaccine with encoding mRNA of virus along the activation of T cells through MHC class-II which elicits protection against SARS-Cov-2 through immune cells from the lymph nodes process the mRNA and synthesize specific viral protein antigens so that other immune cells recognize them. In this vaccine that comprising peptide encoding mRNA along activation of T cells by MHC class-II which immunize body against SARS-Cov-2 are also described. Methods of protecting a host against coronavirus infection will be discussed. Our invented vaccine is type of mRNA vaccine (but combined with other components) against SARS-COV-2 is based on a relatively new genetic method that does not require growing the virus in the laboratory. The technique transforms the human body into a 'living laboratory'. Monkeys given our coronavirus vaccine and then deliberately infected were able to fight off the virus, quickly clearing it from their lungs.
Keywords
Vaccine; SARS-Cov-2; Immune; COVID-19
Introduction
The present paper relates to a designed vaccine useful to protect human against infection by SARS-Cov-2 that causes COVID-19.
Coronaviruses are important human and animal pathogens. At the end of 2019, a novel coronavirus was identified as the cause of a cluster of pneumonia cases in Wuhan, China. It rapidly spread, resulting in an epidemic throughout China, with sporadic cases reported globally. From December 2019 till now, the disease, which resulted in more than twenty millions cases with many deaths, was caused by a novel type of coronavirus, termed COVID-19. Patients with COVID-19 usually developed a high fever followed by clinical symptoms of cough and shortness of breath. Full-genome sequencing and phylogenic analysis indicated that the coronavirus that causes COVID-19 is a beta coronavirus in the same subgenus as the severe acute respiratory syndrome (SARS-CoV) virus (as well as several bat coronaviruses), but in a different clade [1]. The apparent structure of the receptor-binding gene region in COVID-19 is very similar to that of the SARS coronavirus, and there is speculation that it will be shown to use the same receptor for cell entry.
Furthermore, it was found that COVID-19 is 96% identical at the whole-genome level to a bat coronavirus [2]. However, there has been no subsequent consensus regarding which treatment, it any, benefited COVID-19 patients during the outbreak. The development of an effective treatment and vaccination strategy for COVID-19 cases will require clarifying the precise mechanisms by which host immune responses control COVID-19 infection.
Cumulative evidence suggests that patients who recovered from COVID-19 possessed specific acquired immunity based on both T and B cells [3]. Immono cell responses on elimination of pulmonary-Infected COVID-19 and important role of innate immunity have been investigated with details on ref [4].
T-cells are a type of lymphocyte (a type of white blood immune cell) that are involved in bringing about an adaptive immune response. T-cells are so-called ‘T-‘cells because they develop in the thymus. There are two main types of T-cells: CD8+‘killer’ or ‘cytotoxic’ T-cells (Tc cells), and CD4+‘helper’ T-cells (Th cells).
CD8+Tc cells can directly kill pathogen-infected cells as well as recruiting other immune cells by cytokine signaling to ensure a robust immune response is carried out. CD8+Tc cells express T-cell receptors (TCRs) which can recognize specific pathogenic (viral) or foreign antigens.
Any cell infected by a pathogen displays the antigen on its cell surface through class I MHC, to which specific CD8+Tc cells can bind via their TCR. This binding activates an immature CD8+Tc cell to enable it to carry out its cytotoxic (cell-killing) role.
CD4+Th cells on the other hand can recruit B-cells (another lymphocyte which produces antibodies) amongst other cells by recognizing antigens displayed by class II MHC molecules binding to their specific TCR. Also, the cytokines released by CD4+Th cells can allow B-cell antibody class switching as well as the activation and proliferation of CD8+Tc cells [5].
Specifically, CD4+T cells were especially responsive to the viral spike glycoprotein – which is the main target for most vaccines and therapies. These Tc cells are recognizing the spike glycoprotein of the SARS-CoV-2 virus – which is the main site of neutralizing antibodies against the virus. The presence of CD4+Th cells in response to both SARS-CoV and SARS-CoV-2 are generally thought to be favorable in disease outcome.
Any successful vaccine requires the activation of CD4+Th cells which stimulate B-cells to produce antibodies. The evidence suggesting a high degree of T-cell activation in response to SARS-CoV-2 infection (and to some degree in healthy non-infected individuals) is promising in that a robust immune response can be mounted either during COVID-19 infection or through preventative means. Of specific note, the recognition of the receptor binding domain (RBD) is thought to be more effective in viral neutralization rather than generic viral recognition and may be the basis of longer-term immunological memory.
A vast majority of COVID-19 patients develop an immune response against SARS-CoV-2 with the activation of T-cells to the spike-protein of the virus. Researches showed that antibodies of SARS-Cov-2 virus remains in body only 2-3 months but memory T cells remain in body at least 11 years. Therefore, we should consider it in any design of vaccine for SARS-Cov-2.
Background of the invention Coronaviruses are a family of host-specific enveloped RNA viruses with a single-stranded positive sense genome. Examples of coronaviruses include, but are not limited to: feline infectious peritonitis (FIPV) and feline enteric coronavirus (FECV) which are specific to felines; canine coronavirus (CCV) which is specific to canines; transmissible gastroenteritis coronavirus (TGEV) which is specific to swine; bovine coronavirus (BCV) which is specific to bovine species; human coronavirus like SARS-Cov-2 which is specific to humans; mouse hepatitis virus (MHV) which is specific to murine species; and infectious bronchitis virus (IBV) which is specific to avian species.
Coronaviruses share common structural features including a spike or S protein. The S protein is a glycoprotein which protrudes from the surface of the virus particle. The S protein mediates the binding of virions to the host cell receptor and is involved in membrane fusion. In addition, it is the target of virus neutralizing antibodies.
Research shows the viruses bind to the ACE2 receptor through their SB (spike domain B) – which appears ‘crown-like’ and gives rise to the name ‘corona’-virus.
Coronaviruses also require TMPRSS2 to prime the SB-glycoprotein to enter cells through endocytosis.
The SB (spike domain B) of β-coronaviruses is also the main site for antibodies to neutralize the viruses. It is also therefore the target for any successful vaccination for COVID-19 to target the S glycoprotein on the surface of SARS-CoV-2
SARS-CoV-2 (causes COVID-19) has a spike-domain (a glycoprotein) that binds to ACE2 receptors in the lungs and interfering with the spike domain could be a potential vaccine target as well as other therapeutic targets [6].
The spike protein (S protein) is a large type I Trans membrane protein ranging from 1,160 amino acids for avian infectious bronchitis virus (IBV) and up to 1,400 amino acids for feline coronavirus (FCoV) (Figure 1). In addition, this protein is highly glycosylated as it contains 21 to 35 N-glycosylation sites. Spike proteins assemble into trimers on the virion surface to form the distinctive "corona", or crown-like appearance. The ectodomain of all Coronavirus spike proteins share the same organization in two domains: a N-terminal domain named S1 that is responsible for receptor binding and a C-terminal named S2 domain responsible for fusion. Coronavirus diversity is reflected in the variable spike proteins (S proteins), which have evolved into forms differing in their receptor interactions and their response to various environmental triggers of virus-cell membrane fusion [7].
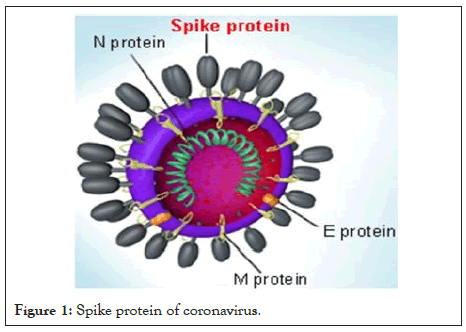
Figure 1: Spike protein of coronavirus.
It's been reported that COVID-19 can infect the human respiratory epithelial cells through interaction with the human ACE2 receptor. Indeed, the recombinant spike protein can bind with recombinant ACE2 protein.
There is a need for a vaccine which can protect against SARS-Cov-2 infection. In particular, there is a need for a vaccine which can be useful to protect human against different coronaviruses, especially SARS-Cov-2 that causes COVID-19.
Detailed Description
According to the present paper, a highly conserved region of the spike protein has been identified which, when presented as a vaccine component or product, is useful as a immunogenic to protect human against SARS-Cov-2 infection. The vaccine of the present in this paper may be used to vaccinate anybody susceptible to infection by virus that is a member of the coronavirus family especially SARS-Cov-2.
As used herein, the term "peptide" is meant to refer to a peptide, polypeptide or protein molecule; a molecule which includes a peptide, polypeptide or protein molecule; or a molecule that contains amino acid residues which are linked by non-peptide bonds.
The virus SARS-CoV-2 uses the spike proteins adorning its outer surface to invade the human cells. SARS-CoV-2 causes the disease COVID-19. A vaccine trains the body’s immune system to recognize some signature viral protein called an antigen. SARS-CoV-2, like other coronaviruses, is named for the crown-like spikes on its surface. There are three proteins on the surface of these viruses: the envelope, membrane and spike, which encapsulate a strand of RNA. This RNA molecule holds the genetic instructions that make up the virus. But viruses do not make their own components. Instead, a SARS-Cov-2 enters into the lung and possibly other respiratory track cells by attaching through to them via its spike protein. Once inside, the viral RNA becomes part of the host cell’s protein production machinery, and produces new copies of viral proteins and RNA which then assemble into thousands of new viruses to spread the disease.
Membrane fusion (viral and cellular) is an essential step when encapsulated viruses enter cells. Fusion of the double lipid layer requires catalysis to overcome a high kinetic barrier, and viral fusion proteins are agents that perform this catalytic function [9]. Although coronavirus uses many different proteins to replicate and invade cells, protein S is the main surface protein that it uses to bind to a receptor - this becomes a bridge to the human cell. After protein S binds to the human cell receptor, the viral membrane fuses with the human cell membrane, allowing the viral genome to enter the human cells and initiate infection (Figure 2) [10].

Figure 2: Architecture and mechanism of TRIM 21 based degradation system. Fc domain is fused to the C-terminus of ABD-targeted peptides. TRIM 21 recognizes Fc domain and tags SARS-Cov-2 for ubiquitin-mediated degradation in the proteasome.
Our invented vaccine is type of mRNA vaccine (but combined with other components) against SARS-COV-2 is based on a relatively new genetic method that does not require growing the virus in the laboratory. The technique transforms the human body into a 'living laboratory', because the whole process is no longer carried out in the laboratory, but directly in the body that received the vaccine.
In a mRNA type vaccine, the technique is actually based on messenger ribonucleic acid (mRNA) fragments, the genetic material that is copied from DNA and encodes proteins. Our invented vaccine encodes the proteins of the new coronavirus, and then injects it into the body. Immune cells from the lymph nodes process the mRNA and synthesize specific viral protein antigens so that other immune cells recognize them. The mRNA is like a software molecule in the body, which then produces viral proteins that can generate an immune response. How the body processes the viral protein is often very different from the way it processes the same protein of the vaccine. Thus, one of the benefits of introducing mRNA into a vaccine is that the body causes the viral protein to behave in exactly the same way that the virus would have directed the host to do.
One way to stop a disease is to block the virus from entering the cells. Vaccines do that by training the body to identify and attack the virus before it can infect healthy human cells (Figure 3).
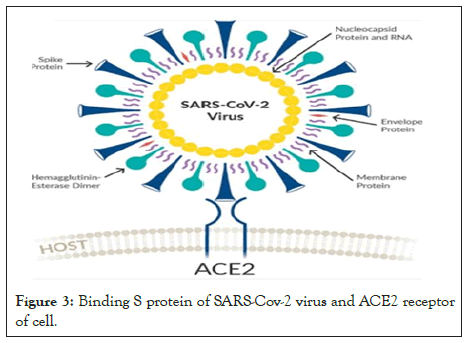
Figure 3: Binding S protein of SARS-Cov-2 virus and ACE2 receptor of cell.
A vaccine is essentially a pure preparation of one or more key components of the virus – such as the envelope, spike or a membrane protein – that is injected in the body to give the immune system a preview of the virus without causing disease. This preview tells the immune system to seek out and attack the virus containing those specific proteins if the real virus ever shows up. However, developing vaccines based on viral proteins takes anywhere from years, such as for the human papilloma virus, to several decades, such as for rotavirus. Protein-based vaccines require mass production of viral proteins in facilities which can guarantee their purity. Growing the viruses and purifying the proteins at medically acceptable pharmaceutical scales can take years. In fact, for some of recent epidemics, such as AIDS, Zika and Ebola, to date there are no effective vaccines.
The present vaccine producing method relates to designed of ACE2-derived peptides which both target the viral spike protein receptor binding domain (RBD) and recruit E3 ubiquitin ligases for subsequent intracellular degradation of SARS-CoV-2 in the proteasome. The engineered peptide fusions demonstrate robust RBD degradation capabilities in human cells. Experimental results identify an optimal peptide variant that can mediate robust degradation of the RBD fused to a stable super folder-green fluorescent protein (sfGFP) in human cells [9].
Numerous previous works have attempted to redirect E3 ubiquitin ligases by replacing their natural protein binding domains with those targeting specific proteins [11]. In 2014 reprogrammed the substrate specificity of a modular human E3 ubiquitin ligase called CHIP (carboxyl-terminus of Hsc70-interacting protein) by replacing its natural substrate-binding domain with designer binding proteins to generate optimized “ubiquibodies” or uAbs. To engineer a single construct that can mediate SARS-CoV-2 degradation without the need for trans expression of TRIM21, the RBD-binding proteins to the CHIPΔTPR modified E3 ubiquitin ligase domain were fused (Figure 4). After cotransfection in HEK293T cells with the RBD-sfGFP complex, the 23-mer (A2N) mutant peptide maintained equivalent levels of degradation between the TRIM21 and CHIPΔTPR fusion architecture were observed, and was more potent than that of sACE2, sACE2v2.4, and the original 23mer (Figure 4) [12].
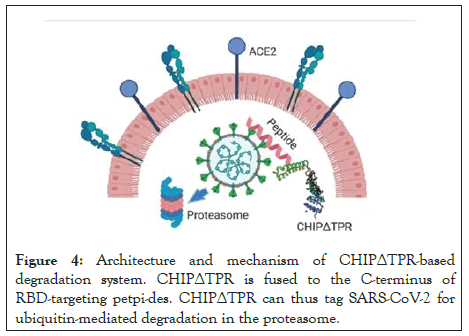
Figure 4: Architecture and mechanism of CHIPΔTPR-based degradation system. CHIPΔTPR is fused to the C-terminus of RBD-targeting petpi-des. CHIPΔTPR can thus tag SARS-CoV-2 for ubiquitin-mediated degradation in the proteasome.
In this invention we used an engineered human ACE2 receptor sequence to potently bind to the SARS-CoV-2 RBD. Present invention uses an optimized peptide variant that enables robust degradation of RBD-sfGFP complexes in human cells, both in Trans and in cis with human E3 ubiquitin ligases. This mutant peptide not only maintains high binding affinity to the RBD, but is also predicted to bind to divergent RBD sequences in the case of future mutation as SARS-CoV-2 continues on its evolutionary trajectory. The peptide will avoid binding to integrin α5β1, thus preventing interference of critical cellular pathways.
Please note, both the peptide and E3 ubiquitin ligase components have been engineered from endogenous human proteins. In terms of in vivo delivery as RNA or recombinant protein, Cas13d has an open reading frame (ORF) of nearly 1000 amino acids, not including the guide RNAs needed for interference. The entire peptide-CHIPΔTPR ORF consists of just over 200 amino acids, which can be readily synthesized as a peptide or be efficiently packaged for delivery in a lipid nanoparticle or adeno-associated virus (AAV) (Figure 5).
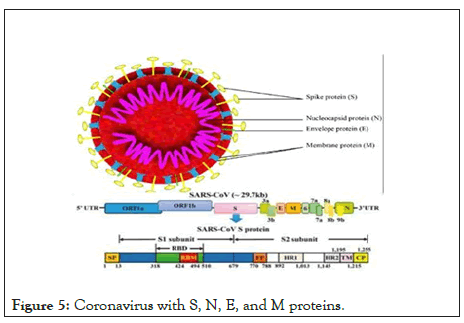
Figure 5: Coronavirus with S, N, E, and M proteins.
Similarly, the peptide-CHIPΔTPR complex can be utilized as a viral interceptor extracellular, as it can competitively bind the RBD to prevent entry via ACE2. In the case that SARS-CoV-2 still infects the cell while boon to peptide-CHIPΔTPR, the virus would be immediately tagged for degradation in the proteasome [9].
As mentioned before, in present method we encode our engineered peptide with mRNA of SARS-Cov 2.
Helper T cells become activated when they are presented with peptide antigens by MHC class II molecules, which are expressed on the surface of antigen-presenting cells (APCs). Once activated, they divide rapidly and secrete cytokines that regulate or assist the immune response. Our engineered peptide may be encoded with MHC class II molecules to activate T cells (especially CD4+). We encode it through MHC Class II YCILEPRSG that presents 14-18 amino acid peptides (Figure 6).
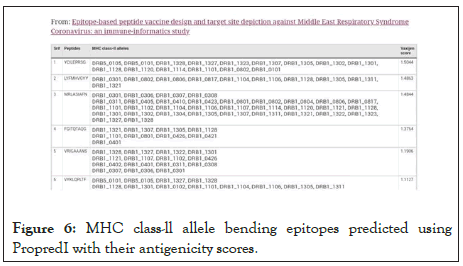
Figure 6: MHC class-ll allele bending epitopes predicted using PropredI with their antigenicity scores.
The main function of major histocompatibility complex (MHC) class II molecules is to present processed antigens, which are derived primarily from exogenous sources, to CD4+ T-lymphocytes. MHC class II molecules thereby are critical for the initiation of the antigen-specific immune response.
Polypeptides or peptides according to the present method may further comprise additional flanking sequences from coronavirus or flanking sequences designed as a consensus sequence of the flanking sequences of corresponding regions from different coronaviruses.
Vaccines according to the present method can be employed to vaccinate human against infection by coronaviruses or at least to prevent the clinical symptoms associated with such infections. Such vaccines will provide protection against multiple coronaviruses (especially SARS-Cov-2) and cross species protection. Vaccines may be produced which are either protein-based or nucleic acid-based. In both cases, the vaccinated human is exposed to an immunogenic peptide or polypeptide. A protective immune response is elicited which is sufficient to protect human against coronavirus.
Vaccines according to the present method can be either: a) compositions which comprise a peptide or polypeptide encoding mRNA along activation of T cells by MHC class-II; or b) compositions which comprise a nucleic acid molecule that includes a nucleotide sequence which encodes a peptide or polypeptide encoding mRNA along activation of T cells by MHC class-II. In both types of vaccines, the peptide or polypeptide is not a complete S protein and it elicits a protective immune response in a human. In protein based, peptides or polypeptides produced using standard techniques including recombinant mRNA techniques for protein production or by peptide synthesis. In preferred embodiments, peptide or polypeptides used in subunit vaccines according to the present invention are produced by recombinant mRNA methodology [13-16].
One having ordinary skill in the art can, using well known techniques, insert DNA molecules into a commercially available expression vector for use in well-known expression systems.
Recombinant vaccines may be used in combination with other vaccines. Further, the genetic material which encodes the peptide or polypeptide may further comprise additional coding sequences which encode other peptide sequences (mRNA along activation of T cells by MHC class-II) capable of eliciting an immunogenic response against coronavirus or another pathogen.
Both subunit and recombinant vaccines may be formulated following accepted convention using buffers, stabilizers, preservative, solubilizes and compositions used to facilitate sustained release. Generally, additives for is tonicity can include sodium chloride, dextrose, manifold, sorbitol and lactose. Stabilizers include gelatin and albumin. Adjuvants such as aluminum or magnesium hydroxide may be employed. Vaccines may be maintained in solution or, in some cases, particularly recombinant vaccines, lyophilized. Lyophilized vaccine may be stored conveniently and combined with sterile solution before administration. Determination of optimum dosage for each parameter may be made by routine methods.
Test
Monkeys given our coronavirus vaccine and then deliberately infected were able to fight off the virus, quickly clearing it from their lungs. We used adult monkeys because in human there is big difference between immune responses of SARS-Cov-2 between adults and children [4]. Please note even immune responses of SARS-Cov-2 between males and females in human are different. Therefore, we don't conclude our vaccine will have same effect as monkeys on human. Other hand, presently the most problem for testing vaccines of SARS-Cov-2 isn't producing antibodies because almost all of them produce it. Main problem is how long this antibody will remain in body because researches show it may disappear after 2-3 months. Therefore, the most important point is to wait 6 months then deliberately infect them again and see if their body is still immune against SARA-Cov-2 either by antibodies or T cells. Therefore, we should test same monkeys after 6 months, and then we can conclude effect of T cell production by MHC class II molecules. Please note if it even shows success after 6 months, it will be for monkeys not human. We still need same types of test for human (Figure 7).
Figure 7: Monkeys given our coronavirus vaccine and then deliberately infected were able to fight off the virus, quickly clearing it from their lungs. We used adult monkeys because in human there is big difference between immune responses of SARS-Cov-2 between adults and children. We should test same monkeys after 6 months, and then we can conclude effect of T cell production by MHC class II molecules.
Discussion
Scientists are pursuing many different strategies to develop new therapeutics against SARS-CoV-2. One area of interest is developing antibodies that bind to and inactivate viral proteins such as the spike protein, which coronaviruses use to enter human cells. A related approach uses small protein fragments called peptides instead of antibodies. Please note that antibodies remain in bodies of recovered patients only 2-3 months that is useless.
The present research relates to a method of protecting a human from infection by a coronavirus (especially SARS-Cov-2) comprising administering an amount of peptides effective to elicit a protective immune response.
In present paper our purpose is setting out to engineer peptides that could strongly bind to the spike protein inside cells, and to use these peptides to trigger the cells to break down the viral proteins.
The idea in this invention is to have the peptides recruit naturally occurring proteins called E3 ubiquitin ligases, which can mark proteins for destruction when cells no longer need them.
All of coronaviruses trigger antibody and T cell responses in infected patients. However, antibody levels appear to wane faster than T cells. COVID-19 antibodies dropped below the limit of detection within 2 to 3 months, whereas SARS-Cov-specific memory T cells have been detected even 11 years after SARS recovered patient.
Helper T cells become activated when they are presented with peptide antigens by MHC class II molecules, which are expressed on the surface of antigen presenting cells (APCs). Once activated, they divide rapidly and secrete cytokines that regulate or assist the immune response.
SARS-CoV-2 (causes COVID-19) has a spike-domain (a glycoprotein) that binds to ACE2 receptors in the lungs and interfering with the spike domain will be used as a vaccine target as well as other therapeutic targets.
In the present paper is used a novel antiviral platform, through design of ACE2-derived peptides which both target the viral spike protein receptor binding domain (RBD) and recruit E3 ubiquitin ligases for subsequent intracellular degradation of SARS-CoV-2 in the proteasome. Our engineered peptide fusions demonstrate robust RBD degradation capabilities in human cells.
The present paper also described a method of protecting human from infection by a SARS-Cov-2 comprising administering an amount of peptides effective to elicit a protective immune response. The peptides administered in the method comprise a conserved domain which can bind an S protein along activation of T cells through MHC class II molecules.
In the present method we also encode our engineered peptide with mRNA of SARS-Cov-2. The mRNA (encoding Ribonucleic Acid) vaccine encodes a stable perfused form (the form before being fused to the cell membrane of the host cell) of Spike protein (S) [8]. Protein complex S is required for membrane fusion and host cell infection and has been the target of vaccines against SARS-Cov-2.
The present paper also relates to a method of protecting a human from infection by a coronavirus (especially SARS-Cov-2) comprising administering an engineered peptide encoding RNA of Coronavirus and encoding MHC class II molecules for activation of T cells to elicit a protective immune response.
Conclusion
1. An engineered peptide with ability of activation of T cells comprising encoding mRNA along MHC class-II encoding for activation of T cells of a SARS-Cov-2 or an immunogenic fragment or derivative thereof; said peptide or polypeptide having less than a complete amino acid sequence of said S protein.
2. A vaccine comprising a pharmaceutically acceptable carrier or diluent and a peptide or polypeptide comprising encoding mRNA along MHC class-II of a SARS-Cov-2 or an immunogenic fragment or derivative thereof; said peptide or polypeptide having less than a complete amino acid sequence of said S protein.
3. A nucleic acid molecule comprises a nucleotide sequence that encodes a peptide or polypeptide comprising a mRNA encoding along MHC class-II encoding of a SARS-Cov-2 or an immunogenic fragment or derivative thereof; said peptide or polypeptide having less than a complete amino acid sequence of said S protein.
4. A recombinant vaccine comprising a nucleic acid molecule, said nucleic acid molecule comprising a nucleotide sequence that encodes a peptide or polypeptide comprising encoding mRNA along MHC class-SARS-Cov-2 II of a or an immunogenic fragment or derivative thereof; said peptide or polypeptide having less than a complete amino acid sequence of said S protein.
5. A method of protecting human against coronavirus comprising administering a peptide or polypeptide with encoding mRNA along MHC class-II of a SARS-Cov-2 or an immunogenic fragment or derivative thereof; said peptide or polypeptide having less than a complete amino acid sequence of said S protein.
6. A method of protecting a human against SARS-Cov 2 comprising administering a nucleic acid molecule comprising a nucleotide sequence that encodes a peptide or polypeptide with encoding mRNA along MHC class-II of a SARS-Cov 2 or an immunogenic fragment or derivative thereof; said peptide or polypeptide having less than a complete amino acid sequence of said S protein.
REFERENCES
- Gorbalenya AE, Baker SC, Baric RS. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020; 5: 536-544.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579(7798): 270-273.
- Zhao J, Zhao J, Van Rooijen N, Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-cov-infected mice. PLoS Pathog. 2009; 5(10): e1000636.
- E. Farshi, B. Kasmapour, A. Arad. "Investigation of immune cells on elimination of pulmonary-Infected COVID-19 and important role of innate immunity, phagocytes," Rev Med Virol. 2020; e2158.
- Uzhachenko RV, Shanker A. CD8+ T lymphocyte and NK cell network: circuitry in the cytotoxic domain of immunity. Front Immunol. 2019; 10: 1906.
- Brouwer P, Caniels T, van Straten K, Snitselaar J, Aldon Y, Bangaru S, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. bioRxiv. 2020.
- Bosch BJ, Van der Zee R, De Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003; 77(16): 8801-8811.
- Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, et al. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell. 2001; 7(3): 593-602.
- Belouzard S, Miller TJ, Klepfer S, Reed AP, Jones EV. Patent: Universal Coronavirus Vaccine 2012.
- Finnegan CM, Berg W, Lewis GK, DeVico AL. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J Virol. 2001; 75(22): 11096-11105.
- Delos SE, Gilbert JM, White JM. The central proline of an internal viral fusion peptide serves two important roles. J Virol. 2000; 74(4): 1686-1693.
- Chatterjee P, Ponnapati M, Jacobson JM. Targeted intracellular degradation of SARS-cov-2 RBD via computationally-optimized peptide fusions. bioRxiv. 2020.
- E. V. Jones, Patent: Universal Coronavirus Vaccine (1993) WO1993023421A1.
- Portnoff AD, Stephens EA, Varner JD, DeLisa MP. Ubiquibodies, synthetic E3 ubiquitin ligases endowed with unnatural substrate specificity for targeted protein silencing. J Biol Chem. 2014; 289(11): 7844-55.
- Bosch BJ, Van der Zee R, De Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003; 77(16): 8801-8811.
- Farshi E. Patent: "Arad Coronavirus Vaccine for SARS-Cov-2" Filed as US patent, 2020.
Citation: Farshi E (2020) Peptide-mRNA Vaccine for SARS-Cov-2. J Vaccines Vaccin. S4:002. DOI: 10.35248/2157-7560.20.S4.002
Copyright: © 2020 Farshi E. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

