PMC/PubMed Indexed Articles
Indexed In
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Scimago
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 15, Issue 6
Nitazoxanide Activating keap1a/Nrf2 Signaling Pathway is Regulated by Cul3 of Criataria plicata
Wuting Lu, Feixiang Su, Fanhua Yang, Jinhua An, Baoqing Hu, Shaoqing Jian, Gang Yang* and Chungen Wen*Received: 11-May-2024, Manuscript No. JARD-24-25658; Editor assigned: 15-May-2024, Pre QC No. JARD-24-25658 (PQ); Reviewed: 29-May-2024, QC No. JARD-24-25658; Revised: 05-Jun-2024, Manuscript No. JARD-24-25658 (R); Published: 12-Jun-2024, DOI: 10.35248/2155-9546.24.15.887
Abstract
Cul3 regulates the ubiquitination and degradation of Nrf2 in Nrf2/keap1a pathway under normal conditions. In aquatic invertebrates, the related mechanism of activating oxidative stress in mussels was studied by stimulating Cristatra plicata with Nitazoxanide (NTZ). The full-length Cul3 gene of C. plicata, designated CpCul3 was cloned. The comparative sequence analysis of CpCul3 gene and its homologous genes showed that CpCul3 gene was highly similar to the homologous genes of other species in amino acid sequence. The amino acid sequence of CpCul3 is 78%-98% similar to that of other invertebrates. In the phylogenetic tree, CpCul3 gene is more closely related to the Cul3 gene in other bivalves. After NTZ stimulation, the expression level of CpCul3 mRNA increased in hepatopancreas of the C. plicata. The protein expression level of CpCul3 was detected by utilizing CpCul3 specific antibody, and the expression level also increased. The results of subcellular localization of CpCul3 in HEK293T indicated that CpCul3 could co-locate with mitochondria, lysosomes, and endoplasmic reticulum, and the colocalization with mitochondria is more obvious. The results indicated that CpCul3 may plays an important role in regulating oxidative stress. The CoIP experiment proved that CpCul3 could promote the ubiquitination degradation of CpNrf2. Lastly, CpNrf2 could be degraded by K48-mediated ubiquitination.
Keywords
Cristatra plicata; Cul3; keap1a/Nrf2; Oxidative stress
Introduction
NTZ is a nitrothiazole benzamide derivative (2-acetyloxy-N-5-nitro- 2-thiazolyl), and Jean Francois Rossignol first used it as an insect repellent against liver nematodes and intestinal repair in 1975, has antibacterial and antiviral effects and can inhibit glioma growth by elevated protein levels of ING1, LC3, and SQSTM1 in mouse [1-4]. Nevertheless, excessive use of NTZ can indeed cause oxidative stress damage to cells and the organism. For instance, the increased mitochondrial ROS levels were detected with the extension of NTZ treatment in MGHU3 cells, and the excessive production of ROS is mainly caused by NTZ-mediated mitosis initiation and lysosomal degradation in the later stage [5]. The drug can also induce ROS overproduction, and increased the levels of ROS, CAT, and GSH after the embryos of zebrafish is exposed to NTZ, but inhibited the activity of SOD and CAT [6].
Oxidative stress refers to the excessive production of Reactive Oxygen Species (ROS) and Reactive Nitrogen Species (RNS) in organism while the body is subjected to various harmful stimuli, the dynamic imbalance of the oxidation system and anti-oxidation system leads to tissue damage [7]. Keap1a/Nrf2 signaling pathway plays an important role in oxidative stress, which is activated by non- biological factors such as heavy metal pollution, organic pollutants, drugs, and biological factors such as bacteria, viruses, toxic algae, parasites [6,8-13]. Under physiological conditions, Nuclear Factor E2-related factor 2 (Nrf2) binds to Kelch-like epichlorohydrin- related protein 1 (Keap1) and undergoes ubiquitination system degradation [14]. Under oxidative stress or in the presence of Nrf2 activators, the structure of Nrf2 and Keap1 complex changes, and Nrf2 protein escapes the inhibitory effect of Keap1, and the synthesized Nrf2 accumulates in cytoplasm and enters into nucleus [15], forming heterodimers with the sMaf protein (MafF/MafG/ MafK) in nucleus [16]. The heterodimer of Nrf2/sMaf regulates the Antioxidant Response Element (ARE) and activates the body’s antioxidant network system, enhancing cellular antioxidant capacity and protecting cells from oxidative damage [17].
Protein ubiquitination is one of the most important post-translational protein modifications, which plays an important role in maintaining intracellular homeostasis and maintains normal cellular physiology, and many cellular functions [18,19]. Human cells express eight different cullins, CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, CUL7, and CUL9, which have evolutionarily conserved Cullin homologous domains at C-terminus responsible for the interaction between cullin and RBX1 or RBX2 [20]. The Cullin3 (Cul3) ubiquitin ligase family consists of three components: the ring finger protein RBX1, the Cul3 scaffold, and the Bricka bruc/Tramwork/Road complex (BTB) protein, which serves as a connection between Cul3 and substrates. The human genome encodes approximately 180 BTB proteins, which means a broad ubiquitination signal and substrate library [21]. Keap1 (Kelch ECH associated protein 1) is the first substrate recognition protein reported to bind to Cullin3 in mammalian thin robes, belonging to the BTB Kelch protein family KLHL protein, which binds to Cullin3 through BTB domain and is responsible for binding to substrates through the Ke1ch domain. Classic substrates include NRF2, IKKβ, p62, and so on [22-24].
Cristaria plicata has strong water filtration ability, which is suitable for habitat, with advantages such as fast pearl formation and strong adaptability, and is an important economic species in aquaculture production in China. The shell is utilized as a button, shell carving serves as a raw material for craftsmanship, and is also used as a mother of pearl. The mussel is consumed as a product, shell powder is designated as feed for livestock and poultry, and is a freshwater pearl industry variety [25,26]. The heavy metals, pesticides, and algal toxins in water environment, affect the survival of mussels. The Keap1/Nrf2 signaling pathway is considered to be the main defense mechanism against cellular oxidative stress. During individual growth, exogenous stimuli can disrupt the cellular oxidative pathway and trigger this signaling pathway [27].
In the present study, we focused on the function of C. plicata CpCul3 and its regulating effect on Keap1-Nrf2/ARE signal pathway. After NTZ stimulation, the mRNA and protein expression levels of CpCul3 were significantly increased in hepatopancreas of the mussel. The subcellular localization results of CpCul3 in HEK293T indicated that it could co-localize with mitochondria, lysosomes, and endoplasmic reticulum. Moreover, CpCul3 could also promote the ubiquitination degradation of CpNrf2, and the promoter activity of CpCul3 is regulated by CpNrf2 and CpMafk.
Materials and Methods
Animals and cells
Healthy Cristaria plicata is collected from Anhui Shuiyun Environmental Protection Co., Ltd in Wuhu, China. The mussels were acclimated for one week at 25°C-30°C with filtered, aerated freshwater. HEK293T cells were grown at 37°C and 5% CO2 with DMEM (C3103-0500, VivaCell) supplemented with 10% fetal bovine serum (C04001-500, VivaCell) and 1% penicillin- streptomycin (Sangon Biotech, China).
Immune stimulation and sample collection
Nitazoxanide (99%, CAS: 55981-09-4) was obtained from Yuanye Biotechnology Co. Ltd (Shanghai, China). 18 mussels were randomly divided into two groups, one group was exposed to 0.3 mg/L NTZ, and another was in equal DMSO. The hepatopancreas tissue was used to extract total RNA or total protein from each group at 4 days poststimulation. The hemolymph from two groups was stained with a Reactive Oxygen Species Assay Kit (Beyotime, S0033S) and was analyzed by a BD LSRFortessa flow cytometer.
cDNA cloning and sequence analysis of CpCul3
The full-length cDNA fragments of CpCul3 were amplified by nested PCR according to the previous report, the primers were designed according to the transcriptome database of our lab [28]. Multiple sequence alignment was constructed with Clustalx from EMBL (https://www.ebi.ac.uk/Tools/msa/clustalo/). The protein structure was predicted with SMART (http://smart.embl-heidelberg.de/). I-TASSER online software (https://seq2fun.dcmb.med.umich.edu//I-TASSER/) and Swiss-PdbViewer (https://spdbv.unil.ch/) predicted the protein 3D structure. A phylogenic tree was constructed with the MEGA 11 software using the neighbor-joining method based on an amino acid alignment. Cul3 sequences from other species were obtained from NCBI (https://www.ncbi.nlm.nih.gov/).
dsRNAi assay
The primers T7- CpCul3-F and T7- CpCul3-R with T7 promoter sequence were used to amplify CpCul3 fragment. Then, it was used to synthesize dsRNA of CpCul3 (604 up). The corresponding control GFP (657 bp) dsRNA was synthesized with the primers T7- EGFP-F and T7-EGFP-R from the pEGFP-C1 vector. The dsRNA was first injected into the mussels at 2 µg of dsRNA per gram of body weight with a microliter syringe through adductor muscle. The mussels were repeatedly injected the equal dsRNA 24 hrs later. The injection volume of the dsRNA solution was adjusted to 100 µL with RNA-free water. The hepatopancreas of each group were collected for the subsequent experiment at 24 hrs after the second injection.
Gene expression profile analysis
The relative expression levels of CpCul3 and antioxidant-related genes in hepatopancreas of the mussels were determined SYBR Green fluorescent RT-qPCR (Vazyme, Nanjing, China) on a CFX96 Real-Time System (Bio-Rad) with specific primers. PCR conditions were 95°C for 2 min, followed by 40 cycles at 95°C for 5 sec, 58°C for 30 sec, and 72°C for 30 sec, and then melting from 65°C to 95°C. The β-actin gene was utilized as an internal control gene of cDNA standardization. The acquired data was analyzed by the 2-ΔΔCt method. Three independent experiments were carried out and the results were expressed as the mean ± SD.
Subcellular localization
HEK-293T cells were passed into confocal culture dishes, after 12- 24 hrs, cells were transfected with indicated plasmids. At 36 hrs post-transfection, the cells were washed twice with PBS and were stained with Mito-Tracker, ER Tracker Red Kit, or Lyso-Tracker Red (Beyotime). Subsequently, Hoechst 33258 solution (Beyotime, China) was applied to stain the nucleus. The treated cells were photographed under a laser scanning confocal microscope (Leica SPE).
Dual-luciferase report gene assays
For investigating the influence of CpNrf2: MafK on CpCul3 gene expression, dual-luciferase report gene assays were executed. The approximate 1 × 105 HEK293T cells seeded in 24 well plates were grown to 70%-80% confluent and transiently co-transfected with 250 ng pGL3-CpCul3 plasm, 50 ng pRL-TK reporter plasmid (Promega) and 250 ng pCDNA3.1-CpNrf2 or pCDNA3.1- CpMafk. To detect the role of bZIP domain on the activity of CpCul3 promoter, the p3xFLAG-14-bZIP plasmid was transfected to cells. Cells transfected with pcDNA3.1 or p3xFLAG-14 plasmid were used as control. Luciferase activity was measured by the Dual- Luciferase Reporter Assay System (Promega). Data was obtained from three independent experimental samples in triplicate.
Immunoblotting
Cells or fresh hepatopancreas tissues were lysed in cold RIPA buffer added with protease inhibitor (20124ES03, Yeasen) in advance. The lysates were centrifuged at max speed (14000 rpm) for 15 min at 4°C. Protein concentration was determined by the BCA protein assay kit (C503021-0501, Sangon). An equal amount of protein (30 µg-50 µg) was analyzed using 10% SDS-PAGE gel and transferred onto PVDF filter membrane. The membrane was blocked with 5% nonfat milk in Tris-Buffered Saline (TBS) (20 mM Tris-HCl, pH 8.0, 150 mM NaCl) for 1-1.5 hrs then incubated with the rabbit anti-CpCul3 (1:4000) overnight at 4°C. After three washes with TBST, the PVDF membranes were incubated with Mouse Anti- Rabbit IgG LCS (A25022, Abbkine). The proteins were detected by the ECL method on the Bio-Rad imaging system.
Co-IP
HEK293T cells were seeded in 10 cm dishes (1 × 107 cells/dish) for 12-24 hrs to form a monolayer of cells that is 70% to 90% confluent. The cells were transfected with indicated plasmids. At 36 hrs post-transfection, the medium was removed carefully, and the cell was washed twice with ice-cold PBS. The cells were then lysed with Cell lysis buffer (P0013, Beyotime) for 30-40 min at 4°C. Lysates were centrifuged at 14,000 rpm for 15 min. The supernatant was incubated with control IgG or Anti-GFP tag nanobody-coated Agarose beads (KTSM1301, AlpaLifeBio). Wash beads 4 times with washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA). Eluting immunoprecipitated proteins from the beads adding 100 µL of 2 × SDS loading buffer and boiling the beads in a thermal block set at 98°C for 10 min. The resin mixture was centrifuged at 2500 × gm for 5 min. The supernatant was evaluated by SDS-PAGE.
Immunohistochemistry.
The tissues of hepatopancreas were fixed in 4% paraformaldehyde and were embedded in paraffin blocks. Then the paraffin was cut into 5 µm. The antigen was restored with a Citric acid antigen repair solution (G1202-250 ML, ServiceBio). The sections were incubated in 0.5% Triton X-100 in PBS for 60 min at RT (Room Temperature). The non-specific signal was blocked using 3% BSA for 1 hr in 0.5% Triton X-100. Hepatopancreas samples were stained with CpCul3 antibody overnight at 4°C and were incubated with 488 fluorescent second antibody. The slides were incubated with an organic spontaneous fluorescence quencher and mounted with a mounting medium. Finally, the slides were imaged with confocal microscopy and were evaluated with fluorescence intensity.
CpCul3 antibody production
The last 597 bp of CpCul3 Open Reading Frame (ORF) sequence was amplified and was ligated into the expression vector pET-28a(+) with 6xHis-tag. The valid recombinant plasmid was extracted and transformed into E. coli BL21 (DE3) Chemically competent cell (TSC-E05, Tsingke, China). A positive colony was induced by Isopropyl-Β-D-Thiogalactose (IPTG) and the protein was purified by Ni-NTA resin. The purified protein was concentrated and quantified with a BCA protein assay kit (Thermo Fisher, USA). To acquire the anti-CpCul3 Ab, that was mixed 0.5 ml of purified CpCul3 protein (1 mg/ml) with an equal volume of Freund’s Complete Adjuvant (FCA) to immunize the New Zealand white rabbit. Half of the antigen was used with Freund’s incomplete adjuvant in the next three immunizations. The serum was tested after the third immunization. Rabbits were sacrificed to obtain the antiserum 7 days after the fourth immunization. Hereafter, the Ab was purified by using a protein A resin column. To prove the effectiveness of the Ab, Western blotting was utilized to detect CpCul3 protein in hepatopancreas through anti-CpCul3, and a single band at the position of 90 kDa for the target was found.
Statistical analysis
The statistical analysis was performed using Student’s t-test or one- way Analysis of Variance (ANOVA), and the data were presented as mean ± SD from 3 independent experiments. Significance was accepted at P<0.05 (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Results
Complementary DNA cloning and bioinformatics analysis of Cul3
The full-length CpCul3 Complementary DNA (cDNA) was cloned from hepatopancreas. The CpCul3 contained a 237 bp 5′ UTR, an 878 bp 3′ UTR, and an ORF. The ORF encoded an 88.7 kDa protein comprising 762 amino acids. Similar to other vertebrates and invertebrates, CpCul3 protein also contained SCOP-d1ldja2, CULLIN, and Nedd8 domains (Figure 1). Additionally, the lengths of ORFs were the same (Figures 1A and 1D). As predicted by the SWISS model, CpCul3 shared high similarity with the tertiary structure of Danio rerio Cul3 (Figure 1B). To further explore the evolutionary relationship of Cul3, the amino acid sequence was used to construct a phylogenetic tree. The mammalian Cul3 clustered into a clade with the homologs in reptiles, gastropods, arthropods, bony fish, and bivalves. The clustering of CpCul3 with its homologs in other various species indicates their close evolutionary relationship (Figure 1C).
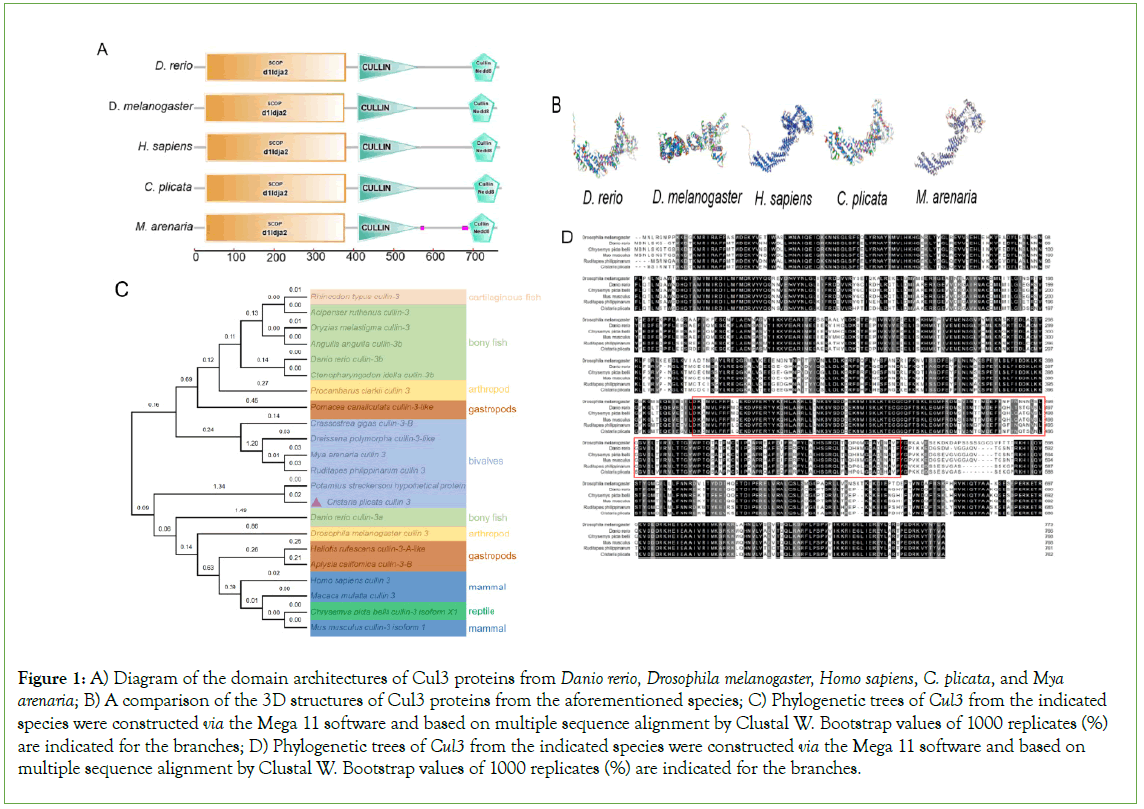
Figure 1: A) Diagram of the domain architectures of Cul3 proteins from Danio rerio, Drosophila melanogaster, Homo sapiens, C. plicata, and Mya arenaria; B) A comparison of the 3D structures of Cul3 proteins from the aforementioned species; C) Phylogenetic trees of Cul3 from the indicated species were constructed via the Mega 11 software and based on multiple sequence alignment by Clustal W. Bootstrap values of 1000 replicates (%) are indicated for the branches; D) Phylogenetic trees of Cul3 from the indicated species were constructed via the Mega 11 software and based on multiple sequence alignment by Clustal W. Bootstrap values of 1000 replicates (%) are indicated for the branches.
Tissue-specific expression of CpCul3
CpCul3 was expressed in all tested mussel tissues, including hepatopancreas and kidney. To further determine the protein expression level of CpCul3 in various tissues of mussels, CpCul3 proteins and synthesized Rabbit polyclonal antibody against CpCul3 was constructed. Western blot assay suggested that CpCul3 was highly expressed in hepatopancreas, kidney, and muscle tissue, while the expression was lower in hemolymph (Figure 2).
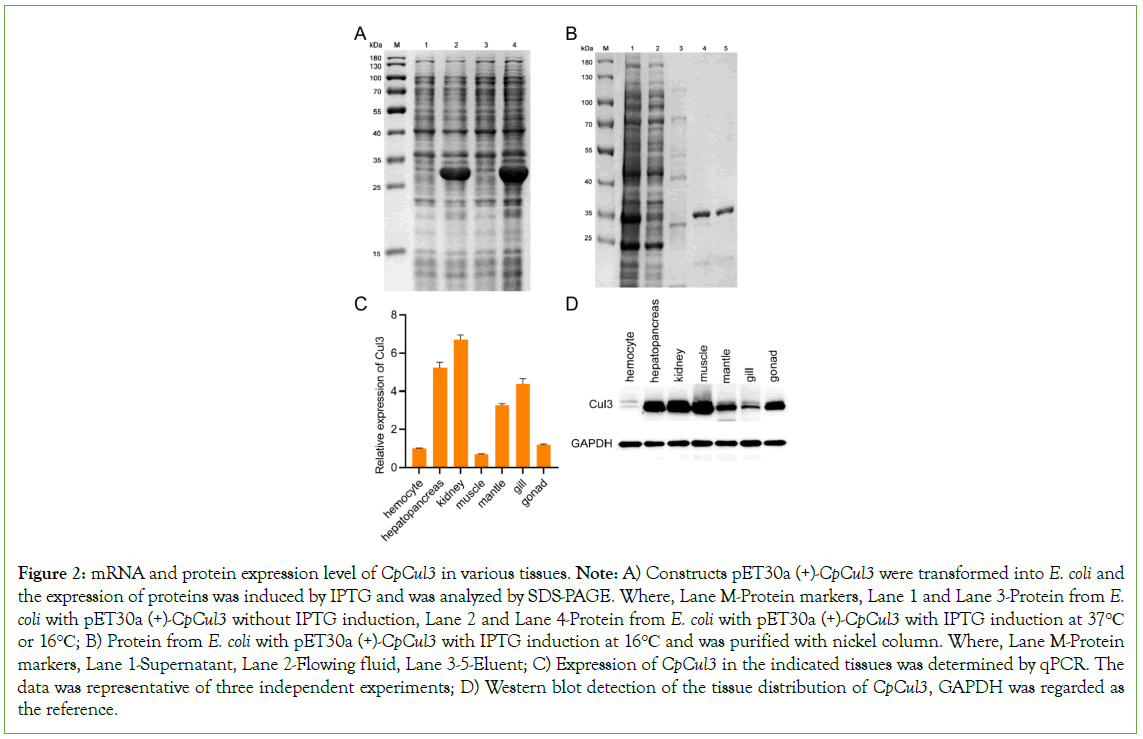
Figure 2: mRNA and protein expression level of CpCul3 in various tissues. Note: A) Constructs pET30a (+)-CpCul3 were transformed into E. coli and the expression of proteins was induced by IPTG and was analyzed by SDS-PAGE. Where, Lane M-Protein markers, Lane 1 and Lane 3-Protein from E. coli with pET30a (+)-CpCul3 without IPTG induction, Lane 2 and Lane 4-Protein from E. coli with pET30a (+)-CpCul3 with IPTG induction at 37°C or 16°C; B) Protein from E. coli with pET30a (+)-CpCul3 with IPTG induction at 16°C and was purified with nickel column. Where, Lane M-Protein markers, Lane 1-Supernatant, Lane 2-Flowing fluid, Lane 3-5-Eluent; C) Expression of CpCul3 in the indicated tissues was determined by qPCR. The data was representative of three independent experiments; D) Western blot detection of the tissue distribution of CpCul3, GAPDH was regarded as the reference.
Participation of CpCul3 in the immune response
The reactive oxygen species in hemolymph increased approximately three times after NTZ exposure (Figure 3A). In hepatopancreas, the expression of CpCul3 was significantly upregulated at 12 hrs, 36 hrs, and 96 hrs after NTZ stimulation (Figure 3B). Meanwhile, CpCul3 mRNA levels were significantly induced in hepatopancreas after NTZ stimulation at 96 hrs (Figure 3C). CpCul3 protein expression was significant in hepatopancreas cytoplasm. In addition, the fluorescence intensity in NTZ exposure group was stronger than that in the control group (Figures 3D and 3E).
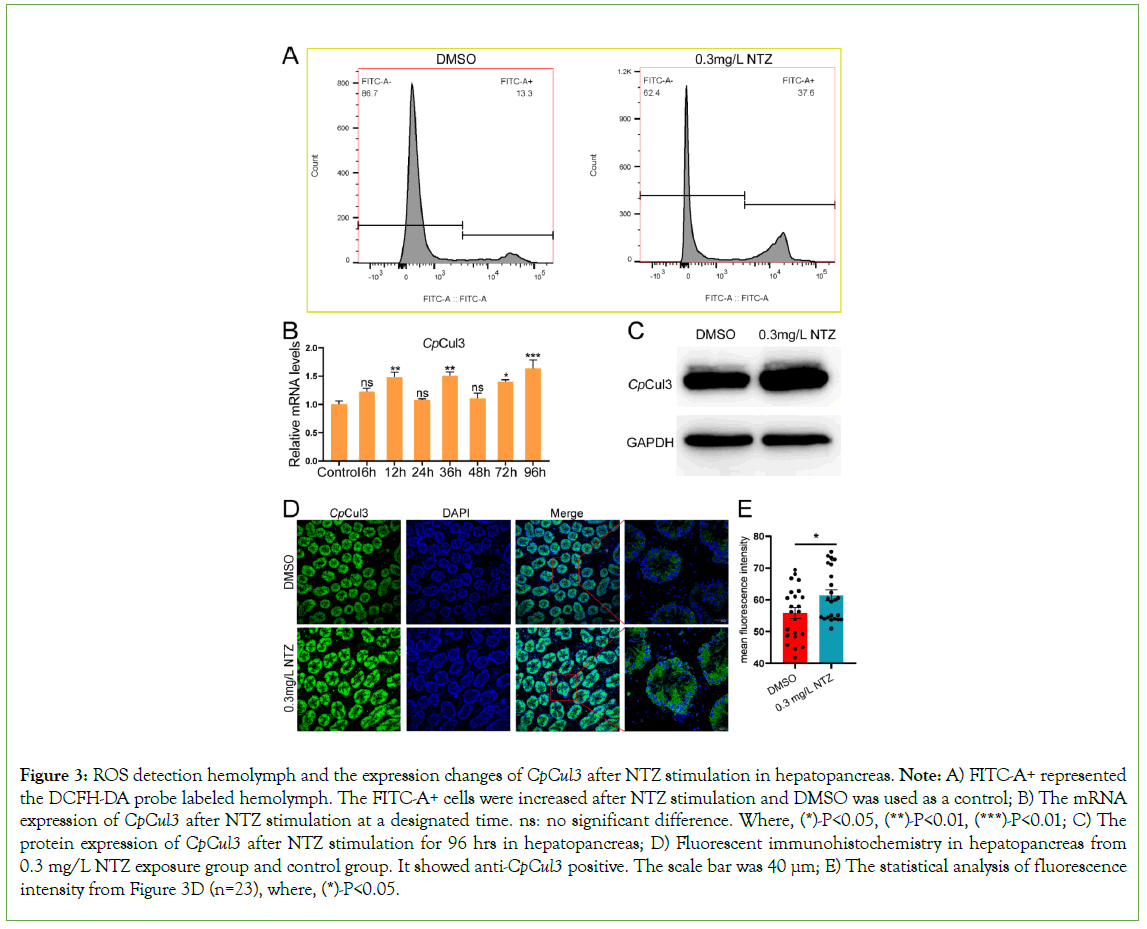
Figure 3: ROS detection hemolymph and the expression changes of CpCul3 after NTZ stimulation in hepatopancreas. Note: A) FITC-A+ represented the DCFH-DA probe labeled hemolymph. The FITC-A+ cells were increased after NTZ stimulation and DMSO was used as a control; B) The mRNA expression of CpCul3 after NTZ stimulation at a designated time. ns: no significant difference. Where, (*)-P<0.05, (**)-P<0.01, (***)-P<0.01; C) The protein expression of CpCul3 after NTZ stimulation for 96 hrs in hepatopancreas; D) Fluorescent immunohistochemistry in hepatopancreas from 0.3 mg/L NTZ exposure group and control group. It showed anti-CpCul3 positive. The scale bar was 40 µm; E) The statistical analysis of fluorescence intensity from Figure 3D (n=23), where, (*)-P<0.05.
Subcellular localization of CpCul3 in HEK293T cells
HEK293T cells were used to investigate the subcellular localization of CpCul3 proteins. As shown in Figure 4, CpCul3-GFP fusion protein was localized in cytoplasm, and had a colocalization with mitochondria, lysosomes, endoplasmic reticulum (Figures 4A-4C).
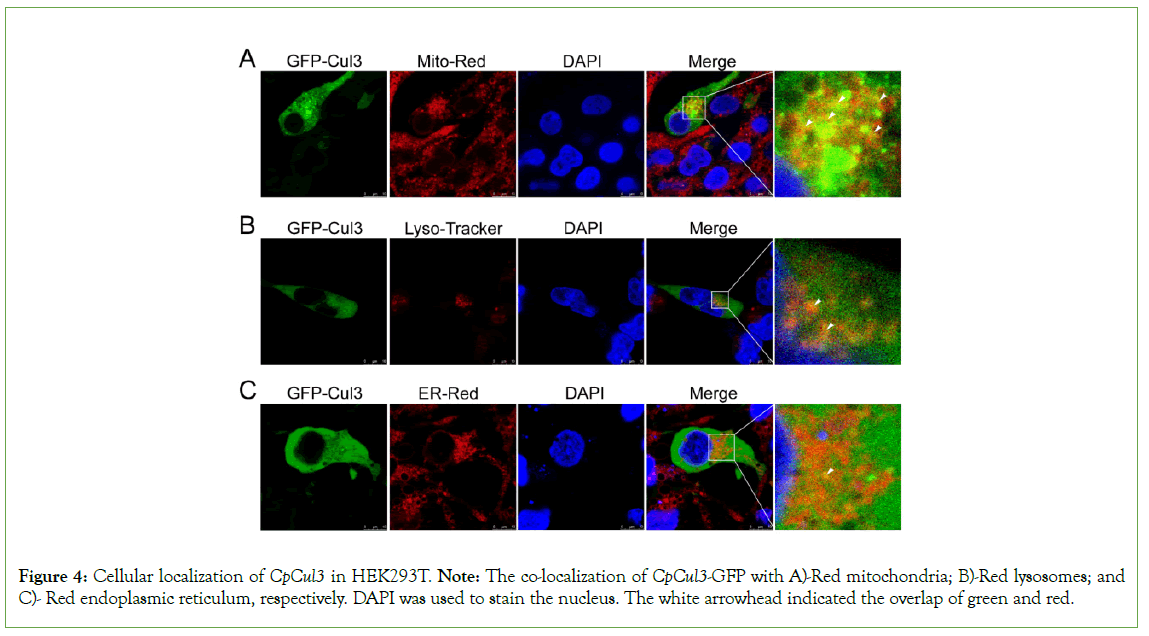
Figure 4: Cellular localization of CpCul3 in HEK293T. Note: The co-localization of CpCul3-GFP with A)-Red mitochondria; B)-Red lysosomes; and C)- Red endoplasmic reticulum, respectively. DAPI was used to stain the nucleus. The white arrowhead indicated the overlap of green and red.
Interaction of CpCul3 with CpRBX1 and Cpkeap1a
CpCul3 interacts with CpRBX1 and Cpkeap1a, respectively the subcellular localization results in HEK293T indicated that CpCul3 had localization with CpRBX1 and Cpkeap1a. Co-IP assays showed that CpCul3 could interact with Cpkeap1a and CpRBX1 (Figure 5).
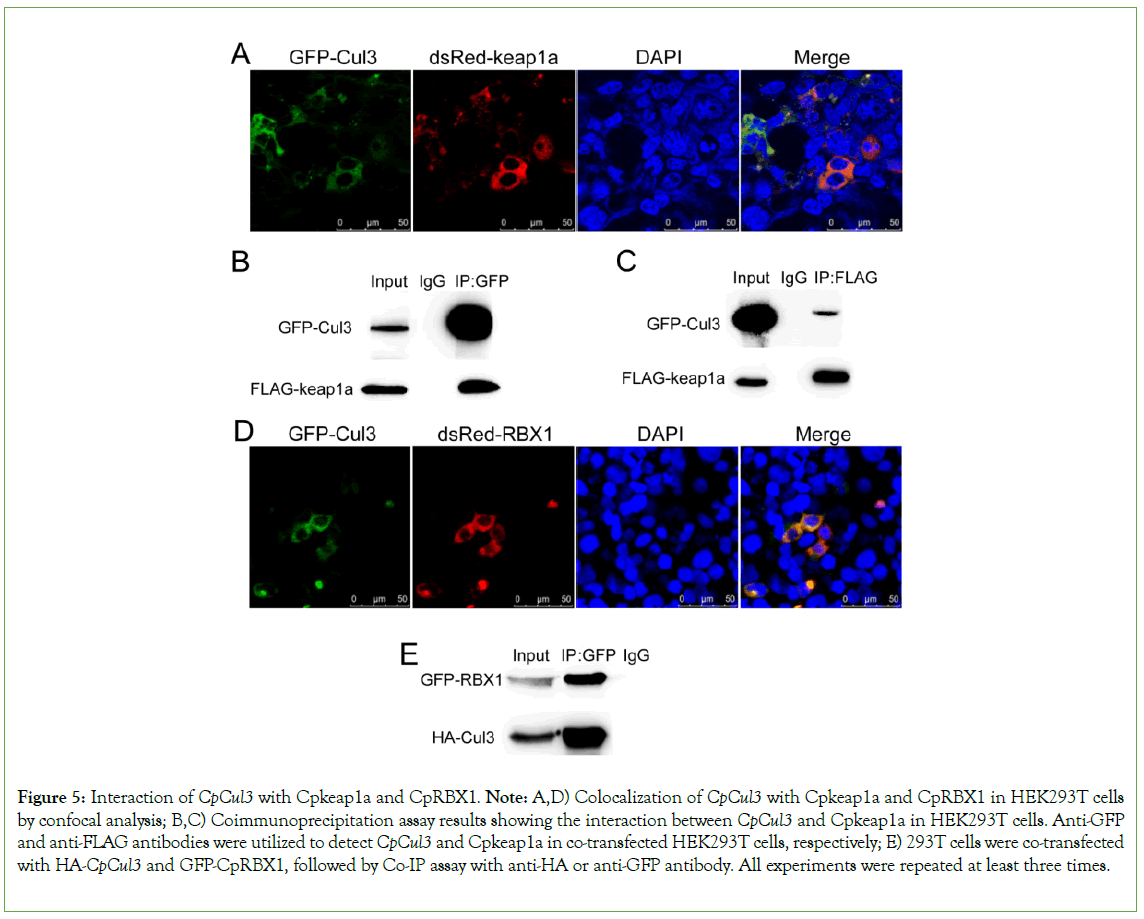
Figure 5: Interaction of CpCul3 with Cpkeap1a and CpRBX1. Note: A,D) Colocalization of CpCul3 with Cpkeap1a and CpRBX1 in HEK293T cells by confocal analysis; B,C) Coimmunoprecipitation assay results showing the interaction between CpCul3 and Cpkeap1a in HEK293T cells. Anti-GFP and anti-FLAG antibodies were utilized to detect CpCul3 and Cpkeap1a in co-transfected HEK293T cells, respectively; E) 293T cells were co-transfected with HA-CpCul3 and GFP-CpRBX1, followed by Co-IP assay with anti-HA or anti-GFP antibody. All experiments were repeated at least three times.
Regulation of CpCul3 by Nrf2/Mafk pathway
A 2512 bp CpCul3 promoter sequence was cloned and had Nrf2, MafK, and MakG binding sites. In CpNrf2 or CpMafk-transfected HEK293T cells, CpCul3 promoter activation was inhibited after NTZ stimulation (Figure 6A). Nrf2 interacts with Mafk through its bZIP domain [29]. In CpNrf2 and CpMafk co-transfected HEK293T cells, CpCul3 promoter activation was inhibited, which were similar to bZIP and CpMafk co-transfected HEK293T cells (Figure 6B). Afterwards, plasmid-protein complexes containing CpMafk fusion proteins and CpCul3 pro-Luc had slower migration rate that was more slowly with the increase of protein concentration (Figure 6C). The results indicated that CpNrf2/Mafk could bind to CpCul3 promoter.
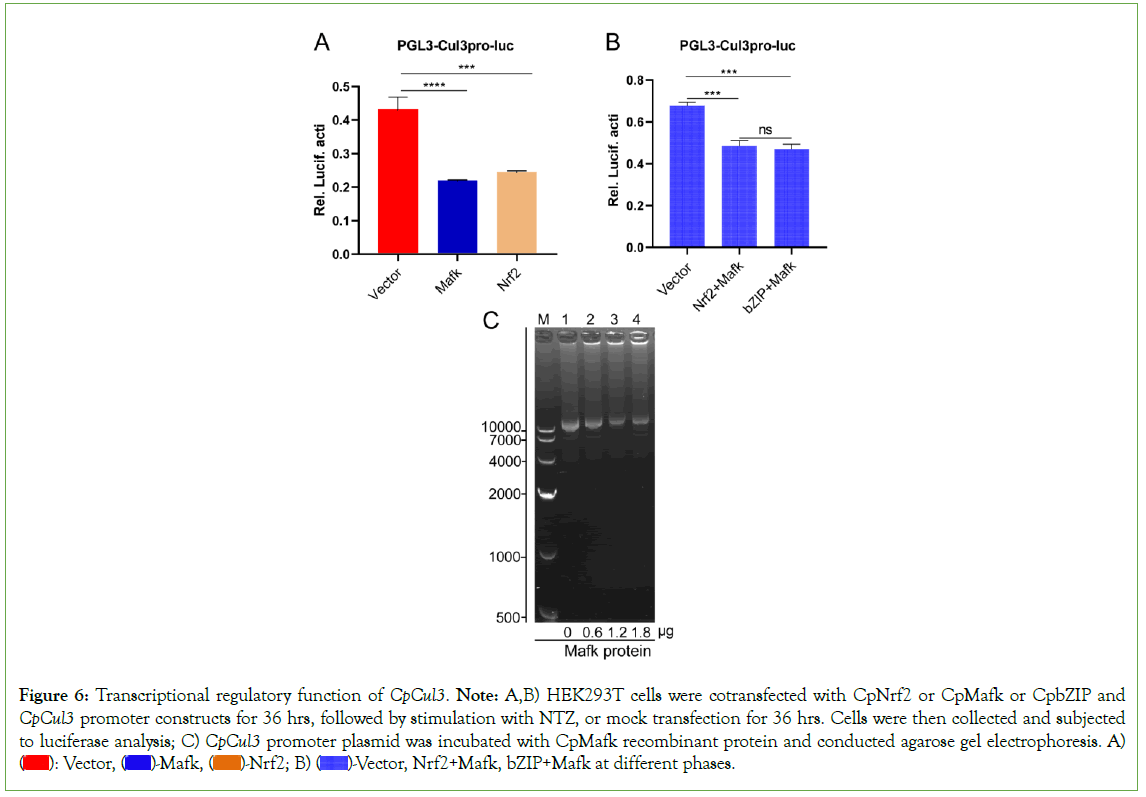
Figure 6: Transcriptional regulatory function of CpCul3. Note: A,B) HEK293T cells were cotransfected with CpNrf2 or CpMafk or CpbZIP and CpCul3 promoter constructs for 36 hrs, followed by stimulation with NTZ, or mock transfection for 36 hrs. Cells were then collected and subjected to luciferase analysis; C) CpCul3 promoter plasmid was incubated with CpMafk recombinant protein and conducted agarose gel electrophoresis. A)  Vector, Nrf2+Mafk, bZIP+Mafk at different phases.
Vector, Nrf2+Mafk, bZIP+Mafk at different phases.
The expression of antioxidant genes were regulated by CpCul3
While CpCul3 was disturbed with double-strand RNA, The expression of CpCul3 was significantly inhibited. The expression level of CAT, GST, and SOD mRNA decreased, while Nrf2, keap1a, and NQO1 relatively increased. The expression of antioxidant genes (CAT, GPX1, Hmox1, NQO1, NFE2L2, PRDX1, SOD1, and SOD2) obviously reduced after the overexpression of CpCul3 in HEK293T cells (Figure 7).
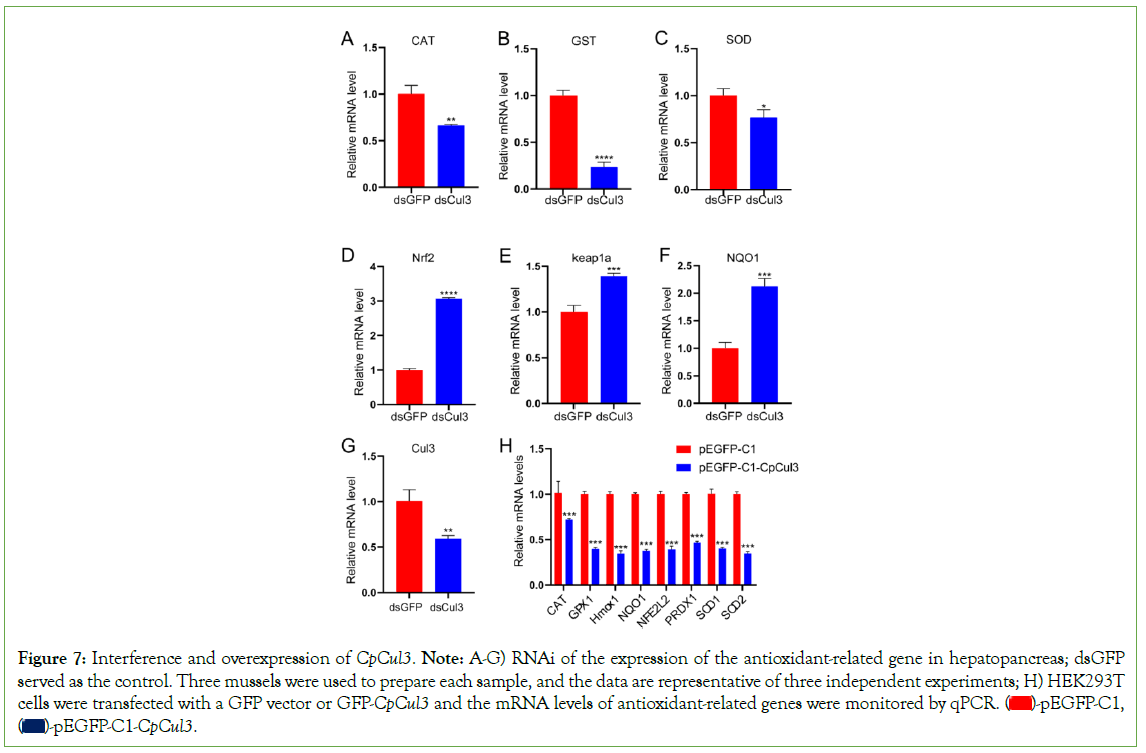
Figure 7: Interference and overexpression of CpCul3. Note: A-G) RNAi of the expression of the antioxidant-related gene in hepatopancreas; dsGFP
served as the control. Three mussels were used to prepare each sample, and the data are representative of three independent experiments; H) HEK293T
cells were transfected with a GFP vector or GFP-CpCul3 and the mRNA levels of antioxidant-related genes were monitored by qPCR.  pEGFP-C1,
pEGFP-C1,  .
.
The ubiquitination of CpNrf2
The plasmid FLAG-CpCul3 was co-transfected with the plasmids HA-Ub and GFP-CpNrf2. The result suggested that CpCul3 facilitated the ubiquitination by CpNrf2 (Figure 8A). The plasmid GFP-CpNrf2 was co-transfected with the plasmids HA-Ub or HA- Ub-K48. The results of immunoprecipitation showed that CpNrf2 could be ubiquitinated by K48-mediated ubiquitination (Figure 8B).
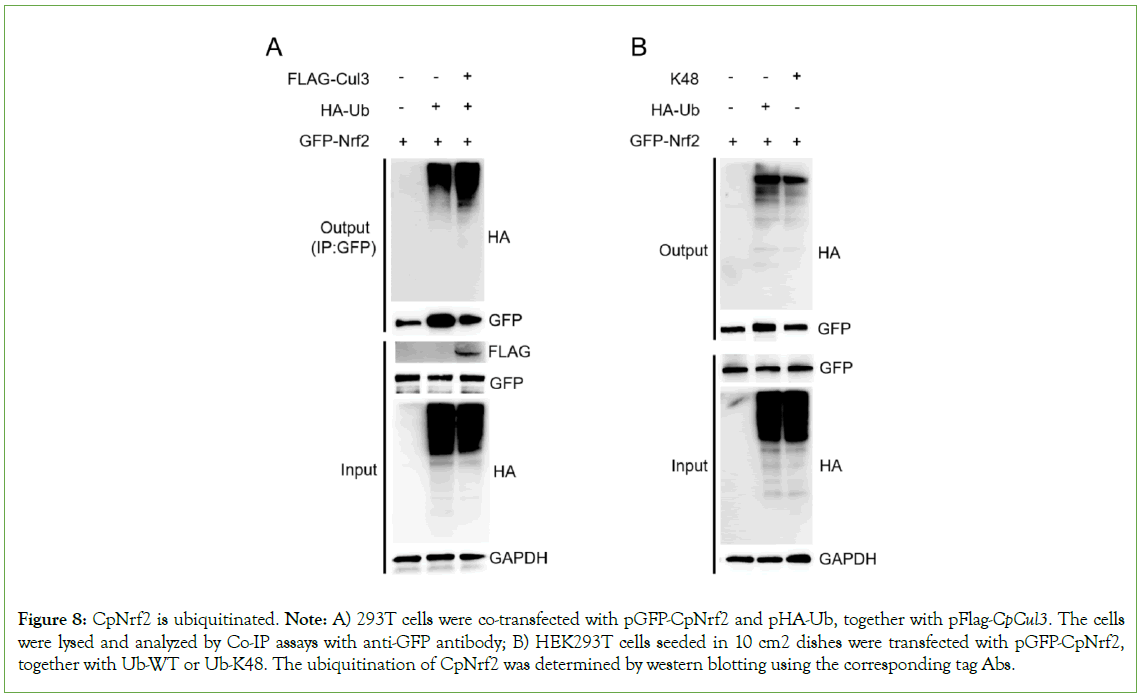
Figure 8: CpNrf2 is ubiquitinated. Note: A) 293T cells were co-transfected with pGFP-CpNrf2 and pHA-Ub, together with pFlag-CpCul3. The cells were lysed and analyzed by Co-IP assays with anti-GFP antibody; B) HEK293T cells seeded in 10 cm2 dishes were transfected with pGFP-CpNrf2, together with Ub-WT or Ub-K48. The ubiquitination of CpNrf2 was determined by western blotting using the corresponding tag Abs.
Discussion
Cul3 is evolutionarily highly conservative, and it contains typical CULLIN and Nedd8 domains [30]. The activity of CRLs (Cullin- RING Ligases) is regulated by post-translational modifications of Nedd8, and these covalent modifications are necessary for the connection between the Cullin protein and the E2 binding enzyme, prompting E3 ligases that recruit ubiquitin molecules to perform ubiquitination modifications on specific substrates [30]. The comparative sequence analysis of CpCul3 gene of C. plicata with its homologs showed that CpCul3 gene had a high similarity with its homolog genes of other species in amino acid sequences. The amino acid sequence of CpCul3 shared 78%-98% similarity with that of other invertebrates. CpCul3 gene also had conservative CULLIN and Nedd8 domains. In the phylogenetic tree, CpCul3 gene was closely related to Cul3 in other bivalves, such as Potamilus streckersoni and Mya arenaria. It was speculated that the function and role of Cul3 might also be conservative in evolution.
In mammals, Cul3 is widely expressed in hepatopancreas, brain, kidney, and ovary [31-34]. The high expression of CpCul3 was detected in hepatopancreas, kidney, and muscle. The tissue of hepatopancreas is an important immune organ of mollusks and plays an important role in immune responses. The relatively high expression levels of CpCul3 in hepatopancreas suggested that it might be involved in the immune process and regulated the expression of immune-related genes. Therefore, the expression level of CpCul3 mRNA in hepatopancreas increased after NTZ stimulation. Meanwhile, CpCul3 antibody was employed to examine the above result under the protein level, which was similar to that of mammals [35]. The above data showed that C. plicata had an evolutionarily conserved Cul3, and might have an immunosuppressive function similar to its mammalian homologs.
In human renal tubular epithelium, Cul3 can co-localize with mitochondria and directly interact with MRPL12 to induce K63- associated ubiquitination of MRPL12, leading to mitochondrial biosynthesis dysfunction [36]. Cul3 is probably involved in the regulation of endosome maturation through ubiquitination, the absence of Cul3 leads to the deformation of endosomes and the defect of endocytosis transport to lysosomes, and Cul3 plays an important role in the later stage of regulating the transport route of lysosomes [37]. The endoplasmic reticulum is the largest membrane-bound organelle in eukaryotes, and it is a continuous structure extending from the nuclear membrane to the plasma membrane [38,39]. The endoplasmic reticulum interacts closely with many organelles, including mitochondria, Golgi apparatus, endosomes, lysosomes, peroxisomes, and plasma membranes, so that lipids and intracellular signals can be transmitted [40]. The endoplasmic reticulum-forming protein Lunapark is ubiquitinated by CRL3klhl12 ubiquitin ligase. Inhibition of Lunapark ubiquitination can lead to neural development defects, and KLHL12-dependent Lunapark ubiquitination is necessary for normal growth and development [41]. The above research shows that Cul3 has a close functional relationship with intracellular organelles. The results of subcellular localization of CpCul3 in HEK293T exhibited that CpCul3 could also co-locate with mitochondria, lysosomes, and endoplasmic reticulum, especially with mitochondria, which is the main structure of cells participating in oxidative stress, indicating that CpCul3 might play an important role in regulating the oxidative stress process.
Cul3, keap1, and RBX1 form a complex to conduct the ubiquitination degradation in cytoplasm, the amino-terminal of Cul3 is connected with keap1 [42], and its carboxyl end interacts with RBX1 [20]. Both subcellular localization and CoIP proved that CpCul3 could interact with Cpkeap1a and CpRBX1, respectively. This indicated that the ubiquitination system was conservative in evolution. Under normal circumstances, Nrf2 is ubiquitinated and degraded in the cytoplasm. The up-regulation of Cul3 can promote the ubiquitination of its substrate, CpCul3 was co-transfected into HEK293T cells with CpNrf2, and the result showed that the ubiquitination level of CpNrf2 also increased [36]. Cul3 ubiquitin ligase can mediate the degradation of K48 polyubiquitination of its substrate CoIP experiment proved that CpNrf2 could indeed undergo K48-linked polyubiquitination in HEK293T cells [43].
Conclusion
As Cul3 can control the ubiquitination of its substrates, we further explored its regulation of the oxidative stress pathway (keap1-Nrf2) through interference and over-expression. Fluorescence quantitative PCR data revealed that the expression levels of antioxidant genes were changed after interfering with CpCul3 in hepatopancreas of C. plicata and overexpressing CpCul3 in HEK293T cells, this result suggested that CpCul3 had a regulatory effect on keap1-Nrf2 pathway, and the double luciferase experiment further verified the negative feedback regulation effect of CpNrf2 on CpCul3 under NTZ stimulation. The above results indicated that CpCul3 played an essential role in keap1-Nrf2 pathway.
In summary, CpCul3 was highly conserved in evolution and widely expressed in various tissues of Cristaria plicata. The CpCul3 mRNA and protein expression levels increased under NTZ stimulation. CpCul3 could promote the ubiquitination of CpNrf2. CpNrf2 and CpMafk could bind to the promoter of Cul3 to regulate the expression of CpCul3.
Acknowledgments
Funding
This research was financially supported by grants (No. 331460697, 13007168, 13007198) from the National Natural Science Foundation of China, the Support Project of the Modern Agricultural Industry Technology System (ZQT20180027), the Scientific and Technological in Jiangxi Province (20160BBF60053, 20192BAB204009), the Jiangxi Agriculture Research System (JXARS-03).
Credit authorship contribution statement
Wuting Lu: Investigation, conceptualization, methodology, formal analysis, writing-original draft; Feixiang Su: Validation, resources; Fanhua Yang: Laser confocal photography; Jinhua An: Software, data curation; Baoqing Hu: Investigation, funding acquisition; Shaoqing Jian: Validation, methodology. Gang Yang: Supervision; Chungen Wen: Writing-review and editing, supervision, funding acquisition.
Conflict of interest
The authors of this article declare no conflict of interest.
References
- Fox LM, Saravolatz LD. Nitazoxanide: A new thiazolide antiparasitic agent. Clin Infect Dis. 2005;40(8):1173-1180.
[Crossref] [Google Scholar] [PubMed]
- Dubreuil L, Houcke I, Mouton Y, Rossignol JF. In vitro evaluation of activities of nitazoxanide and tizoxanide against anaerobes and aerobic organisms. Antimicrob Agents Chemother. 1996;40(10):2266-2270.
[Crossref] [Google Scholar] [PubMed]
- Al-Kuraishy HM, Al-Gareeb AI, Elekhnawy E, Batiha GE. Nitazoxanide and COVID-19: A review. Mol Biol Rep. 2022;49(11):11169-11176.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Shen C, Liu Z, Peng F, Chen X, Yang G, et al. Nitazoxanide, an antiprotozoal drug, inhibits late-stage autophagy and promotes ING1-induced cell cycle arrest in glioblastoma. Cell Death Dis. 2018;9(10):1032.
[Crossref] [Google Scholar] [PubMed]
- Sun H, Ou T, Hu J, Yang Z, Lei Q, Li Y, et al. Nitazoxanide impairs mitophagy flux through ROS-mediated mitophagy initiation and lysosomal dysfunction in bladder cancer. Biochem Pharmacol. 2021;190:114588.
[Crossref] [Google Scholar] [PubMed]
- Lu W, Yang F, Meng Y, An J, Hu B, Jian S, et al. Immunotoxicity and transcriptome analysis of zebrafish embryos exposure to Nitazoxanide. Fish Shellfish Immunol. 2023;141:108977.
[Crossref] [Google Scholar] [PubMed]
- Cai L, Kang YJ. Oxidative stress and diabetic cardiomyopathy: A brief review. Cardiovasc Toxicol. 2001;1(3):181-193.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Liao G, Tu H, Huang Y, Peng T, Xu Y, et al. A protective role of autophagy in Pb-induced developmental neurotoxicity in zebrafish. Chemosphere. 2019;235:1050-1058.
[Crossref] [Google Scholar] [PubMed]
- Wang Q, Gu X, Liu Y, Liu S, Lu W, Wu Y, et al. Insights into the circadian rhythm alterations of the novel PFOS substitutes F-53B and OBS on adult zebrafish. J Hazard Mater. 2023;448:130959.
[Crossref] [Google Scholar] [PubMed]
- Jiang WD, Hu K, Liu Y, Jiang J, Wu P, Zhao J, et al. Dietary myo-inositol modulates immunity through antioxidant activity and the Nrf2 and E2F4/cyclin signalling factors in the head kidney and spleen following infection of juvenile fish with Aeromonas hydrophila. Fish Shellfish Immunol. 2016;49:374-386.
[Crossref] [Google Scholar] [PubMed]
- Singh E, Matada GSP, Abbas N, Dhiwar PS, Ghara A, Das A. Management of COVID-19-induced cytokine storm by Keap1-Nrf2 system: A review. Inflammopharmacology. 2021;29(5):1347-1355.
[Crossref] [Google Scholar] [PubMed]
- Falfushynska H, Horyn O, Osypenko I, Rzymski P, Wejnerowski L, Dziuba MK, et al. Multibiomarker-based assessment of toxicity of central European strains of filamentous cyanobacteria Aphanizomenon gracile and Raphidiopsis raciborskii to zebrafish Danio rerio. Water Res. 2021;194:116923.
[Crossref] [Google Scholar] [PubMed]
- Zhou W, Quan JH, Lee YH, Shin DW, Cha GH. Toxoplasma gondii proliferation require down-regulation of host nox4 expression via activation of PI3 Kinase/Akt signaling pathway. PLoS One. 2013;8(6):e66306.
[Crossref] [Google Scholar] [PubMed]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130-7139.
[Crossref] [Google Scholar] [PubMed]
- Yang Y, Cvekl A. Large Maf transcription factors: Cousins of AP-1 proteins and important regulators of cellular differentiation. Einstein J Biol Med. 2007;23(1):2-11.
[Crossref] [Google Scholar] [PubMed]
- Katsuoka F, Yamamoto M. Small Maf proteins (MafF, MafG, MafK): History, structure and function. Gene. 2016;586(2):197-205.
[Crossref] [Google Scholar] [PubMed]
- Pellegrino S, Ronda L, Annoni C, Contini A, Erba E, Gelmi ML, et al. Molecular insights into dimerization inhibition of c-Maf transcription factor. Biochim Biophys Acta. 2014;1844(12):2108-2115.
[Crossref] [Google Scholar] [PubMed]
- Ciechanover A. The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J 1998;17(24):7151-7160.
[Crossref] [Google Scholar] [PubMed]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425-479.
[Crossref] [Google Scholar] [PubMed]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703-709.
[Crossref] [Google Scholar] [PubMed]
- Chen RH. Cullin 3 and its role in tumorigenesis. Adv Exp Med Biol. 2020;1217:187-210.
[Crossref] [Google Scholar] [PubMed]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477-8486.
[Crossref] [Google Scholar] [PubMed]
- Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKβ. Mol Cell. 2009;36(1):131-140.
[Crossref] [Google Scholar] [PubMed]
- Lee Y, Chou TF, Pittman SK, Keith AL, Razani B, Weihl CC. Keap1/Cullin3 modulates p62/SQSTM1 activity via UBA domain ubiquitination. Cell Rep. 2017;19(1):188-202.
[Crossref] [Google Scholar] [PubMed]
- Lecoeur S, Videmann B, Berny P. Evaluation of metallothionein as a biomarker of single and combined Cd/Cu exposure in Dreissena polymorpha. Environ Res. 2004;94(2):184-191.
[Crossref] [Google Scholar] [PubMed]
- Li D, Pan B, Chen L, Wang Y, Wang T, Wang J. et al. Bioaccumulation and human health risk assessment of trace metals in the freshwater mussel Cristaria plicata in Dongting Lake, China. J Environ Sci. 2021;104:335-350.
[Crossref] [Google Scholar] [PubMed]
- Lushchak VI. Environmentally induced oxidative stress in aquatic animals. Aquat Toxicol. 2011;101(1):13-30.
[Crossref] [Google Scholar] [PubMed]
- Cao X, Lu W, Gang Y, Hu B, Wen C. Prx5 of Cristaria plicata has antioxidant function and is regulated by Nrf2/ARE signaling pathway. Fish Shellfish Immunol. 2023;134.
[Crossref] [Google Scholar] [PubMed]
- Otsuki A, Yamamoto M. Cis-element architecture of Nrf2-sMaf heterodimer binding sites and its relation to diseases. Arch Pharm Res. 2020;43(3):275-285.
[Crossref] [Google Scholar] [PubMed]
- Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7(10):1014-1020.
[Crossref] [Google Scholar] [PubMed]
- Zhao M, Quan Y, Zeng J, Lyu X, Wang H, Lei JH, et al. Cullin3 deficiency shapes tumor microenvironment and promotes cholangiocarcinoma in liver-specific Smad4/Pten mutant mice. Int J Biol Sci. 2021;17(15):4176-4191.
[Crossref] [Google Scholar] [PubMed]
- Morandell J, Schwarz LA, Basilico B, Tasciyan S, Dimchev G, Nicolas A, et al. Cul3 regulates cytoskeleton protein homeostasis and cell migration during a critical window of brain development. Nat Commun. 2021;12(1):3058.
[Crossref] [Google Scholar] [PubMed]
- Maeoka Y, Cornelius RJ, Ferdaus MZ, Sharma A, Nguyen LT, McCormick JA. Cullin 3 mutant causing familial hyperkalemic hypertension lacks normal activity in the kidney. Am J Physiol Renal Physiol. 2022;323(5):F564-F576.
[Crossref] [Google Scholar] [PubMed]
- Li X, Yang KB, Chen W, Mai J, Wu XQ, Sun T, et al. CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy. 2021;17(12): 4323-4340.
[Crossref] [Google Scholar] [PubMed]
- Wu Y, Zhao M, Lin Z. Pyrroloquinoline quinone (PQQ) alleviated sepsis-induced acute liver injury, inflammation, oxidative stress and cell apoptosis by downregulating CUL3 expression. Bioengineered. 2021;12(1):2459-2468.
[Crossref] [Google Scholar] [PubMed]
- Ji X, Yang X, Gu X, Chu L, Sun S, Sun J, et al. CUL3 induces mitochondrial dysfunction via MRPL12 ubiquitination in renal tubular epithelial cells. FEBS J. 2023;290(22):5340-5352.
[Crossref] [Google Scholar] [PubMed]
- Huotari J, Meyer-Schaller N, Hubner M, Stauffer S, Katheder N, Horvath P, et al. Cullin-3 regulates late endosome maturation. Proc Natl Acad Sci USA. 2012;109(3):823-828.
[Crossref] [Google Scholar] [PubMed]
- Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr Opin Cell Biol. 2013;25(4):428-433.
[Crossref] [Google Scholar] [PubMed]
- Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17(2):69-82.
[Crossref] [Google Scholar] [PubMed]
- Friedman JR, Voeltz GK. The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol. 2011;21(12):709-717.
[Crossref] [Google Scholar] [PubMed]
- Yuniati L, Lauriola A, Gerritsen M, Abreu S, Ni E, Tesoriero C, et al. Ubiquitylation of the ER-shaping protein Lunapark via the CRL3(KLHL12) ubiquitin ligase complex. Cell Rep. 2020;31(7):107664.
[Crossref] [Google Scholar] [PubMed]
- Zhao Y, Sun Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr Pharm Des. 2013;19(18):3215-3225.
[Crossref] [Google Scholar] [PubMed]
- Jerabkova K, Sumara I. Cullin 3, a cellular scripter of the non-proteolytic ubiquitin code. Semin Cell Dev Biol. 2019;93:100-110.
[Crossref] [Google Scholar] [PubMed]
Citation: Lu W, Su F, Yang F, An J, Hu B, Jian S, et al (2024) Nitazoxanide Activating keap1a/Nrf2 Signaling Pathway is Regulated by Cul3 of Criataria plicata. J Aquac Res Dev. 15:887.
Copyright: © 2024 Lu W, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.