Indexed In
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2022) Volume 8, Issue 2
Molecular and Morphological Identification of Fish Gut Microbiota at Different Growth Stage of Labeo rohita
Muhammad Waqar Mazhar*Received: 21-Mar-2022, Manuscript No. CMBO-22-15809; Editor assigned: 25-Mar-2022, Pre QC No. CMBO-22-15809 (PQ); Reviewed: 12-Apr-2022, QC No. CMBO-22-15809; Revised: 19-Apr-2022, Manuscript No. CMBO-22-15809 (R); Published: 26-Apr-2022, DOI: 10.35841/2471-2663.22.8.119
Abstract
Introduction: The gut micro-biota plays a significant role in the fish health and pathogenicity. L.rohita is well known farmed fish it also culture with other major carps.
Methodology: The present study was conducted to determine the molecular and morphological identification of gut micro-biota of L.rohita collected from the fish hatchery Faisalabad. Isolation of bacteria was carried out through the culture techniques. The morphological identification of bacterial isolates was determined by the gram staining, biochemical tests TSI and MR-VP test. The molecular identification was done by 16S rRNA techniques.
Results: The results of present research work was showed that the 20 strains of bacterial were isolated from the gut of L.rohita and all these isolates were gram negative and morphological identify on the basis of biochemical tests i.e. TSI and MR-VP test. Biofilm result indicated that six, eight and six isolates showed the weak, moderate and strong biofilm.
Conclusion: The highest and lowest resistance showed against cefadroxil and Levofloxin respectively. The lowest intermediate isolates were found against cefadroxil, polymyxin B and colistin and highest intermediate were found against ceftriaxone. The highest sensitive isolates were found against Levofloxin and lowest sensitive isolate were found in cefadroxil, nitrofurantoin and cefoxitin. On the basis of 16Sr RNA sequencing and phylogenetic relationship among isolates 17 isolate were determined in the present study. Klebsiella pneumonia, Enterobacter cloacae, Pseudomonas oleovorans, Morganella morganii, Citrobacter freundii, Proteus mirabilis, Citrobacter braakii, Enterobacter hormaechei, Psychrobacter sanguinis, Shigella dysenteriae, Citrobacter cronae, Shigella sonnei, Pseudomonas sihuiensis, Proteus mirabilis, Enterobacter hormaechei were found in the intestine of L.rohita.
Keywords
L.rohita; 16Sr RNA; Levofloxin; Cefadroxil; MR-VP test
Introduction
In the previous year, the aquaculture is one of the rapidly increasing food productions in the world [1]. Economically it has great importance for public and private sectors. In the EU, the utilization of fish as food increase consistently [2]. Fish has a great source of protein but it also related to GI track infection in humans. The L.rohita found in Asia and in the world its production was 1.5 million tons in 2012 [3]. It is an herbivore; eurythermal species not survive below 14°C. The fish gut is an open system and contains different microbial population [4]. In recent years’ scientist use term prebiotics reporting as health facilitating bacteria, these bacteria may be helpful in decreasing the infections and seriousness of epidemics [5]. Lactic acid bacteria have prebiotic effect and these bacteria microflora present in GI track of mammals and aquatic animals with no damaging effect. The finfish GI tract and gut microbes play important role in host health [6]. The fish has different types of microflora and quantity affected by microbial population of water, sand, mud, water and their habitat [7]. Enterobacteriaceae are microbes that present in GI track of humans and mammals have not dangerous effect on host health and in some cases it creates diseases under favorable condition [8].
Methodology
Sample collection
T15 samples of Labeo rohita of different growth sages were collected, anesthetized with clove oil 75 ul/l in water. Fish sample were disinfectant with 70% ethanol, dissected with use of scalper and isolated gut packed into bags and store at -20°C.
Isolation of bacteria from fish gut
Stored gut sample was taken and small portion of gut was cut with sharp blade and placed in petri dish, about 1 g was added into LB media and placed into flask in the shaking incubator for overnight to grow. Inoculum was placed into autoclaved MacConkey agar and steaking plates were incubated for overnight at 37°C. Then 50 ul inoculum was used for spreading in media plates with the help of spreader. Different colonies were obtained on plates and steaked on another plate to obtain pure culture and incubate at 37°C to grow.
Biochemical identification
Sample was placed on slide and slide dipped into crystal violet de for 30s and then washed with 70% ethanol and then dipped in the safranin de for 30s and washed with tap water. After drying slides were observed under 100x light microscope.
Triple sugar iron test
Colonies steak on the slant that present in test tube, TSI media was used in slants and incubate at 37°C for overnight. The next day result was calculated and slant color changes due to presence of bacteria and produce H2S gas.
MR-VP test
Single colony picked and mixed in MRVP media present in test tube. Placed in shaking incubator for 48 hrs at 37°C. PH noted by using PH strips. After checking the PH 400 ul 5% alpha-napthol shakenly and add 15 ul of 40% KOH placed in shaking incubator for 1 hr.
Extraction of genomic DNA
Single colony picked and mixed with TSB media in Eppendorf, placed in shaking incubator at 37°C for overnight. Growth were appeared and centrifuge at 10000 rpm for 5 minutes to remove supernatant. The DNA isolation was done by phenol chloroform method. Store DNA at 4°C.
Molecular analysis on genomic DNA
By adding PCR water, Taq buffer (1X), primers (0.4 μM), dNTPs (0.2 Mm) (MgCl2 1.5 Mm) and Taq polymerase (2 units) master mix was prepared. In the PCR tubes equal amount of master mix placed and then equal amount of DNA mixed. After that PCR tubes placed in thermocycler and optimize the profile. After completing 35 cycles the product confirmation was done by 1.5% agarose gel electrophoresis (Tables 1 and 2).
| Sr. No. | Reagents | Master mix | Concentrations |
|---|---|---|---|
| 1 | Buffer (1X buffer+(NH4)2SO4 | 5 | 1X |
| 2 | MgCl2(mM) | 3 | 1.5 |
| 3 | dNTPs(mM) | 1 | 0.2 |
| 4 | Primer µM | 4 | 0.4 |
| 5 | Taq Polymerase (uints) | 0.4 | 2 |
| 6 | DNA template length ng/µl | 1 | 1 |
| 7 | PCR Water µl | 35.6 | |
| Total | 50µl | ||
Table 1: Composition of PCR reagents for the amplification of genomic DNA of isolated gut micro biota from Labeo rohita collected from fish hatchery Faisalabad.
| Sr. No. | Steps in PCR | Stages | Temperature °C | Time | Cycle |
|---|---|---|---|---|---|
| 1 | Denaturation | I | 95 | 30 sec | |
| 2 | Primer annealing | II | 56.5 | 30 sec. | |
| 3 | Primer extension | III | 72 | 1 min | 35 |
| 4 | Final extension | 72 | 10 min | ||
| 5 | Hold temperature | 8 |
Table 2: PCR used for the amplification of genome pattern in 16s RNA marker.
Bio film study
The fresh culture was diluted in 0.2% glucose L.B media by 1:100 ratios and from that diluted culture 125 μl was added in wells of 96 well plate three wells for each sample. Then added the crystal violet and placed for 10 minutes, washed plate for three times and incubate the plate for two hours. After two hours added 30% acetic acid and left for 10 minutes. Checked the results in ELISA reader.
Antibiotic sensitivity test
The sensitivity of bacteria measured by using Hudzicki and Kirby- Bauer method. Different zone appeared on antibiotic agar plates and results were noted.
Results
The total 20 types of bacterial colony were isolated from 15 fish gut samples and grow on MacConkeyagar media. Two types of colonies observed one was sugar fermented and other was non sugar fermented. Due to fermented process sugar fermented bacteria converted into red color while non fermented into brown color (Figures 1 and 2).

Figure 1: The non-sugar fermenter bacterial colony isolate from the gut of labeo.
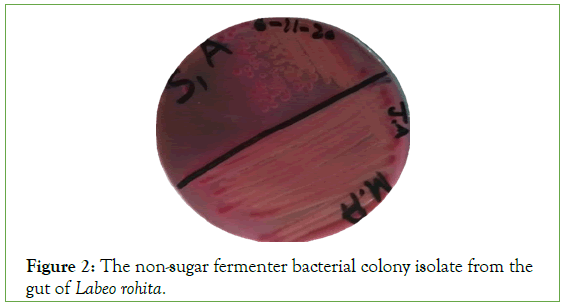
Figure 2: The non-sugar fermenter bacterial colony isolate from the gut of Labeo rohita.
Tripe sugar iron test
The triple sugar iron test on the basis of lactose fermentation and nonlactose fermentation and H2S gas production. Six bacteria showed the altered media color that change red to yellow color but slant change color and produce gas and not H2S gas. Two bacterial isolates changes the butt color red to black and slans remain same and no H2S production (Figure 3 and Table 3).
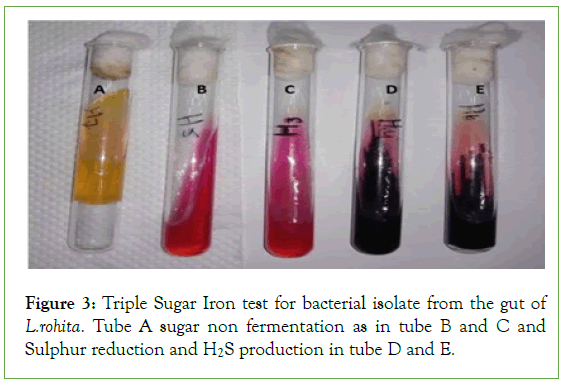
Figure 3: Triple Sugar Iron test for bacterial isolate from the gut of L.rohita. Tube A sugar non fermentation as in tube B and C and Sulphur reduction and H2S production in tube D and E.
| Sr. no | Sample | Butt color | Slant color | Gas | H2S |
|---|---|---|---|---|---|
| 1 | S1-A | Yellow | Yellow | Yes | No |
| 2 | S1-B | Yellow | Red | Yes | No |
| 3 | S2-1 | Yellow | Yellow | Yes | No |
| 4 | S2-2 | Red | Red | No | No |
| 5 | S2-3 | Yellow | Yellow | Yes | No |
| 6 | S3-A | Red | Red | No | No |
| 7 | S3-B | Yellow | Yellow | No | No |
| 8 | S3-3 | Yellow | Red | Yes | No |
| 9 | S3-4 | Black | Red | Yes | Yes |
| 10 | S3-7 | Black | Red | Yes | Yes |
| 11 | S4-1 | Yellow | Yellow | Yes | No |
| 12 | S4-2 | Red | Red | No | No |
| 13 | S5-A | Red | Red | No | No |
| 14 | S5-B | Yellow | Red | Yes | No |
| 15 | S5-C | Yellow | Red | Yes | No |
| 16 | H3 | Red | Red | No | No |
| 17 | H4 | Black | Red | No | No |
| 18 | H5 | Black | Red | No | No |
| 19 | H6 | Black | Red | Yes | Yes |
| 20 | H7 | Yellow | Yellow | Yes | No |
Table 3: Triple Sugar Iron test for bacterial isolate from the gut of L. rohita.
MR-VP test
The results indicated that 13 bacterial strains showed the MR positive while 7 showed negative. On the other hand, 9 samples showed VR positive and 11 showed VR negative (Figure 4 and Table 4).
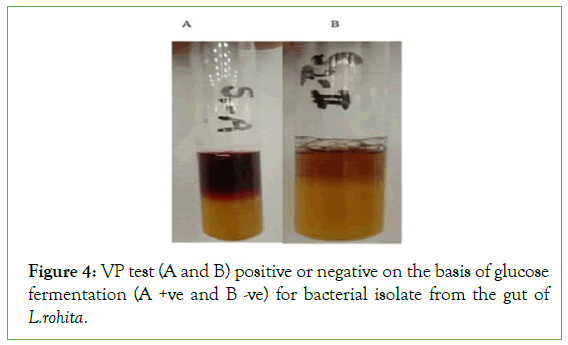
Figure 4: VP test (A and B) positive or negative on the basis of glucose fermentation (A +ve and B -ve) for bacterial isolate from the gut of L.rohita.
| Sr. no | Sample | MR | VP | Biofilm |
|---|---|---|---|---|
| 1 | S1-A | +ve | +ve | M |
| 2 | S1-B | -ve | +ve | M |
| 3 | S2-1 | -ve | +ve | M |
| 4 | S2-2 | -ve | -ve | S |
| 5 | S2-3 | +ve | -ve | W |
| 6 | S3-A | +ve | -ve | W |
| 7 | S3-B | +ve | -ve | W |
| 8 | S3-3 | +ve | -ve | M |
| 9 | S3-4 | +ve | -ve | S |
| 10 | S3-7 | +ve | +ve | S |
| 11 | S4-1 | -ve | +ve | S |
| 12 | S4-2 | -ve | -ve | M |
| 13 | S5-A | +ve | -ve | S |
| 14 | S5-B | +ve | +ve | M |
| 15 | S5-C | +ve | -ve | M |
| 16 | H3 | -ve | +ve | W |
| 17 | H4 | +ve | -ve | W |
| 18 | H5 | +ve | -ve | M |
| 19 | H6 | +ve | +ve | M |
| 20 | H7 | -ve | +ve | W |
Table 4: The MR VP and biofilm test for bacterial isolate from the gut of L.rohita.
Biofilm
The result of biofilm on the basis of ELISA technique S2-2, S3- 4, S3-7, S4-1, S5-A, (six isolate) showed the weak biofilm and S1- A, S1-B, S2-1, S3-3, S4-2, S5-B, S5-C, H5 and H6 (eight isolate) showed the moderate Biofilm while S2-3, S3-A, S3-B, H3, H4 and H7 (six isolates) showed the strong biofilm.
Antibiotic resistance
The result of antibiotic resistance against different antibiotic. The results indicated that S1-A, S1-B, S2-1, S2-2, S2-3, S3-A, S3-B, S3-3, S3-4, S3-7, S4-1, H3, H5, showed the strong antibiotic resistance and S4-2, S5-A, S5-C, H4, H7 showed sensitivity while S5-B and H6 showed the intermediate against amoxicillin clavualanic acid. Same results were obtained against except S5-B which showed the resistance and no bacterial isolate strain showed the intermediate against cefadroxil (Figures 5 and 6 and Table 5).
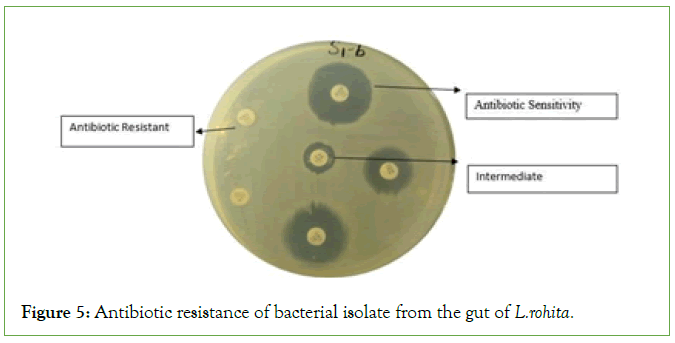
Figure 5: Antibiotic resistance of bacterial isolate from the gut of L.rohita.
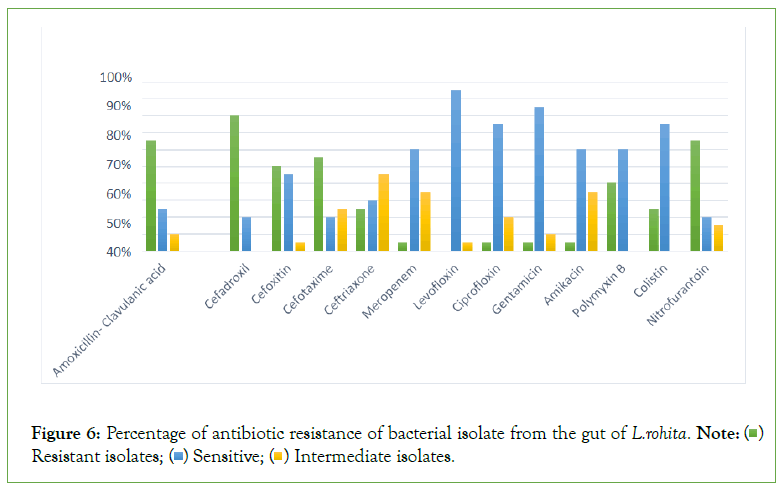
Figure 6: Percentage of antibiotic resistance of bacterial isolate from the gut of L.rohita Note: ( ) Resistant isolates; (
) Resistant isolates; ( ) Sensitive; (
) Sensitive; ( ) Intermediate isolates.
) Intermediate isolates.
| Sr. no | Antibiotics | S1- A | S1- B | S2-1 | S2-2 | S2-3 | S3-A | S3- B | S3-3 | S3-4 | S3-7 | S4-1 | S4-2 | S5- A | S5- B | S5- C | H3 | H4 | H5 | H6 | H7 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Amoxicillin clavulanic acid | R | R | R | R | R | R | R | R | R | R | R | S | S | I | S | R | S | R | I | S |
| 2 | Cefadroxil | R | R | R | R | R | R | R | R | R | R | R | S | S | R | R | R | S | R | R | S |
| 3 | Cefoxitin | S | R | R | S | S | S | R | R | R | S | R | R | S | R | S | I | S | R | R | S |
| 4 | Cefotaxime | R | R | R | R | R | I | S | S | R | I | I | S | R | R | R | R | S | R | I | I |
| 5 | Ceftriaxone | R | I | I | I | R | S | I | I | S | S | I | S | S | I | R | I | R | R | I | S |
| 6 | Meropenem | I | S | I | S | S | S | S | I | S | I | S | S | S | I | S | S | R | I | S | I |
| 7 | Levofloxin | S | S | S | S | S | S | S | S | S | S | S | S | S | S | I | S | S | S | S | S |
| 8 | Ciprofloxin | I | S | S | S | I | R | S | S | S | S | S | S | S | S | I | S | I | S | S | S |
| 9 | Gentamicin | S | S | S | R | S | S | S | S | S | S | S | S | S | S | S | S | I | S | S | I |
| 10 | Amikacin | I | S | S | S | S | S | S | I | S | S | S | S | I | I | I | S | I | S | I | R |
| 11 | Polymyxin B | S | R | R | S | S | R | S | R | S | R | S | S | S | R | R | S | R | S | S | S |
| 12 | Colistin | S | S | S | S | S | R | S | S | S | S | R | S | S | S | S | S | R | R | R | S |
| 13 | Nitrofuranto in | R | S | R | S | I | R | R | R | S | R | R | R | R | S | I | R | R | R | I | R |
Table 5: Antibiotic resistance of 20 isolates from the gut of Labeo rohita.
DNA extraction and molecular analysis
In this study DNA extraction done by organic and lysate DNA extraction method. The concentration of DNA quantifies at 260- 280 nm wavelength by using UV spectrophotometer. Amplification of 16S rRNA genes was carried by using universal primers. PCR product of 20 isolates analyzed by 1.5% agarose gel (Figure 7 and Table 6).

Figure 7: Amplification of 16S rRNA of 20 isolates from gut of L.rohita.
| Sr. No. | Isolates | Assigned taxonomic name | Max score | Similarity | Accession number |
|---|---|---|---|---|---|
| 1 | S1-A | Klebsiella pneumoniae | 968 | 95.62% | NR_117686.1 |
| 2 | S2-B | Enterobacter cloacae | 878 | 97.42% | NR_117679.1 |
| 3 | S2-1 | ||||
| 4 | S2-2 | ||||
| 5 | S2-3 | Pseudomonas oleovorans | 488 | 91.49% | NR_115874.1 |
| 6 | S3-A | Morganella morganii | 1022 | 94.66% | NR_113580.1 |
| 7 | S3-B | Citrobacter freundii | 1048 | 97.26% | NR_113340.1 |
| 8 | S3-3 | Proteus mirabilis | 850 | 92.23% | NR_113344.1 |
| 9 | S3-4 | Citrobacter braakii | 811 | 88.67% | NR_028687.1 |
| 10 | S3-7 | ||||
| 11 | S4-1 | Enterobacter hormaechei | 1122 | 97.00% | NR_126208.1 |
| 12 | S4-2 | Psychrobacter sanguinis | 819 | 89.91% | NR_117833.1 |
| 13 | S5-A | Shigella dysenteriae | 1027 | 95.50% | NR_026332.1 |
| 14 | S5-B | Citrobacter cronae | 1020 | 97.50% | NR_170426.1 |
| 15 | S5-C | Shigella sonnei | 1058 | 98.66% | NR_104826.1 |
| 16 | H3 | Pseudomonas sihuiensis | 893 | 91.02% | NR_148251.1 |
| 17 | H4 | Proteus mirabilis | 830 | 92.29% | NR_113344.1 |
| 18 | H5 | Pseudomonas oleovorans | 1081 | 97.93% | NR_115874.1 |
| 19 | H6 | Citrobacter cronae | 1014 | 95.45% | NR_170426.1 |
| 20 | H7 | Enterobacter hormaechei | 1011 | 98.27% | NR_042154.1 |
Table 6: The assigned taxonomic name and Max score of 20 bacterial strains along with GenBank accession numbers.
Phylogenetic tree
Comparison of 16S rRNA sequence in NCBI GenBank USING BAST program showed the similarities of 20 isolates were 88.67% to 98.27% (Figure 8).
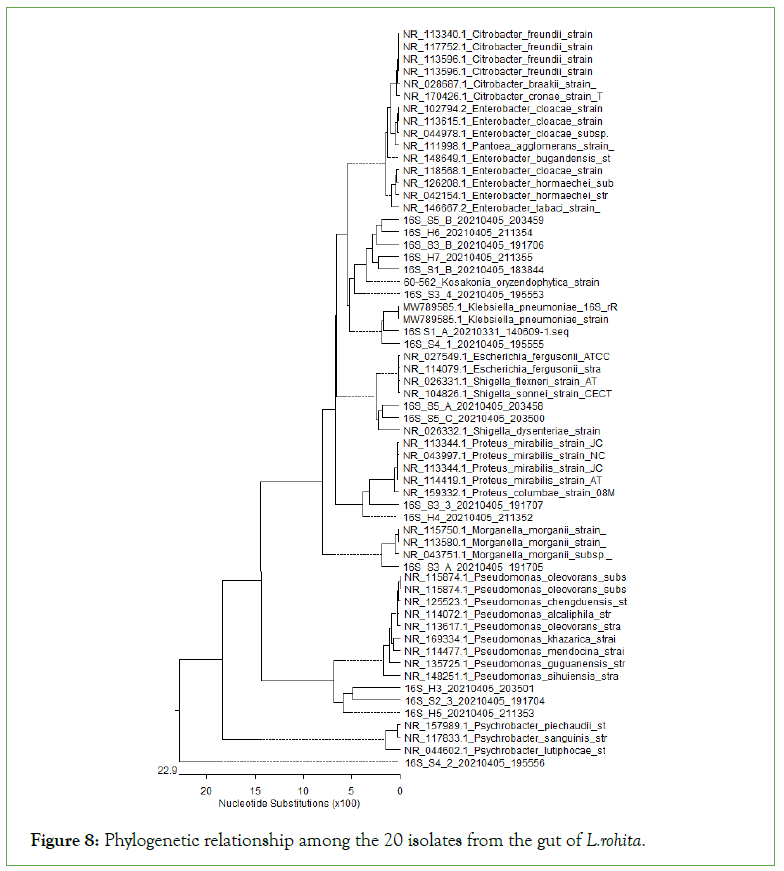
Figure 8: Phylogenetic relationship among the 20 isolates from the gut of L.rohita.
Discussion
Fish is the desire able source of protein and unsaturated fatty acid. Now days, fish lipids have presumed a great nutritional implication due to their protective role against the development of cardiovascular diseases and rheumatoid arthritis [9]. Rahu is commercially important fish and normally feed on planktons, plant matter and decaying vegetation [10].
The fish gut is considered as an open system it contains microbial populations from the aquatic environment through water and food which are populated with bacteria [11]. Micro-biota of fish gut paly a significant role in health and disease [12] Fish gut micro-biota confers various effects to the host this includes size, metabolism, feeding behavior and immune response in the fish It is also necessary to increase the growth of small sized fish and develop the protection of fish against microbial pathogens and gut micro-biota resolve the both issues [13].
Identification was performed considering all morphological biochemical and molecular characters of isolated gut micro-biota. In the present study the 20 bacterial isolates were gram negative these results were correlated with previous studies [14]. Seventeen isolates were isolated from the gastro-intestinal tract and gill of major carps in which ten isolates were gram positive and seven were gram negative. Tathagata et al. found the 50% gram negative and 50% gram positive colonies in the intestinal micro-biota of 8 samples of fish.
On the basis of biochemical test the present study morphological identified the bacterial isolated strains and these were related with these biochemical tests for the purification of the isolate bacterial colonies.
Biofilm production and antibiotic resistance were also determined in the present studies. Several antibiotic resistance strains have been reported by researchers due to the indiscriminate antibiotic use in aquaculture farms [15]. The antibiotic and metal resistance in bacterial population is clearly an environmental phenomenon of natural selection for survival. Bacterial resistance for antibiotics and metals may be due to the presence of R-Plasmid [16]. Ten antibiotics were exposed to check the susceptibility of bacterial isolates while thirteen antibiotics were used to check the sensitivity and resistance of bacterial isolate from the gut of L.rohita [17].
The predominant bacteria found in the intestine of L.rohita included Pseudomonas, Aeromonas, Alcaligens, Citrobacter, Enterobacter, E. coli, Salmonella, Bacillus, Streptococcus, Clostridium, but Klebsiella, Serratia, Proteus, Staphylococcus, Chromobacterium, Flavobacterium, Micrococcus, Corynebacterium, Shigellawere found to be moderate [18]. The results of present study were somewhat correlate with the above study.
Conclusion
The highest and lowest resistance showed against cefadroxil and Levofloxin respectively. The lowest intermediate isolates were found against cefadroxil, polymyxin B and colistin and highest intermediate were found against ceftriaxone. The highest sensitive isolates were found against Levofloxin and lowest sensitive isolate were found in cefadroxil, nitrofurantoin and cefoxitin. On the basis of 16Sr RNA sequencing and phylogenetic relationship among isolates 17 isolate were determined in the present study. Klebsiella pneumonia, Enterobacter cloacae, Pseudomonas oleovorans, Morganella morganii, Citrobacter freundii, Proteus mirabilis, Citrobacter braakii, Enterobacter hormaechei, Psychrobacter sanguinis, Shigella dysenteriae, Citrobacter cronae, Shigella sonnei, Pseudomonas sihuiensis, Proteus mirabilis, Enterobacter hormaechei were found in the intestine of L.rohita.
Conflict of Interest
All authors have no conflict of interest.
Consent for Publication
This study based on research.
Funding
No funds.
Authors Contribution
All authors have contribution in this study.
Acknowledgement
This research study is self-funded.
Availability of Data and Materials
The data associated with a paper is available, the data availability on demand through email contact of co-author.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
REFERENCES
- Edwards P, Zhang W, Belton B, Little DC. Misunderstandings, myths and mantras in aquaculture: its contribution to world food supplies has been systematically over reported. Mar Policy. 2019;106:103547.
- Schmidt O, Padel S, Levidow L. The bio-economy concept and knowledge base in a public goods and farmer perspective. Bio-based Appl Econ. 2012;1(1):47-63.
- Harikrishnan R, Devi G, Van Doan H, Balasundaram C, Arockiaraj J, Jagruthi C. Efficacy of ulvan on immune response and immuno-antioxidant gene modulation in Labeo rohita against columnaris disease. Fish Shellfish Immunol. 2021;117:262-273.
[CrossRef] [GoogleScholar] [PubMed]
- Sullam KE, Essinger SD, Lozupone CA, O'Connor MP, Rosen GL, Knight R, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21(13):3363-3378.
[CrossRef] [GoogleScholar] [PubMed]
- Hu J, Zhang L, Lin W, Tang W, Chan FKL, Ng SC. Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci Technol. 2021;108:187-196.
[CrossRef] [GoogleScholar] [PubMed]
- Ringø E, Løvmo L, Kristiansen M, Bakken Y, Salinas I, Myklebust R, et al. Lactic acid bacteria vs. pathogens in the gastrointestinal tract of fish: A review. Aquac Res. 2010;41(4):451-467.
- Paliaga P, Felja I, Budiša A, Ivančić I. The impact of a fish cannery wastewater discharge on the bacterial community structure and sanitary conditions of marine coastal sediments. Water. 2019;11(12):2566.
- Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome. 2019;7(1):1-15.
[CrossRef] [GoogleScholar] [PubMed]
- Kandemir Ş, Polat N. Seasonal Variation of Total Lipid and Total Fatty Acid in Muscle and Liver of Rainbow Trout (Oncorhynchus mykiss W, 1792) Reared in Derbent Dam Lake. Turkish J Fish Aquat Sci. 2007;7(1).
- Leoni G, Neumann PA, Sumagin R, Denning TL, Nusrat A. Wound repair: Role of immune-epithelial interactions. Mucosal Immunol. 2015;8(5):959-968.
[CrossRef] [GoogleScholar] [PubMed]
- Blancheton JP, Attramadal KJ, Michaud L, d’Orbcastel ER, Vadstein O. Insight into bacterial population in aquaculture systems and its implication. Aquac Eng. 2013;53:30-39.
- Banerjee S, Mukherjee A, Dutta D, Ghosh K. Evaluation of chitinolytic gut microbiota in some carps and optimization of culture conditions for chitinase production by the selected bacteria. J microbiol biotechnol food sci. 2021;2021:12-19.
- Yukgehnaish K, Kumar P, Sivachandran P, Marimuthu K, Arshad A, Paray BA, et al. Gut microbiota metagenomics in aquaculture: Factors influencing gut microbiome and its physiological role in fish. Rev Aquac. 2020;12(3):1903-1927.
- Shabana, Shahid SU, Irfan U. The gut microbiota and its potential role in obesity. Future Microbiol. 2018;13:589-603.
[CrossRef] [GoogleScholar] [PubMed]
- Yuyama KT, Wendt L, Surup F, Kretz R, Chepkirui C, Wittstein K, et al. Cytochalasans Act as Inhibitors of Biofilm Formation of Staphylococcus Aureus. Biomolecules. 2018;8(4):129.
[CrossRef] [GoogleScholar] [PubMed]
- Majumdar RK, Bejjanki SK, Roy D, Shitole S, Saha A, Narayan B. Biochemical and microbial characterization of Ngari and Hentaak-traditional fermented fish products of India. J Food Sci Technol. 2015;52(12):8284-8291.
[CrossRef] [GoogleScholar] [PubMed]
- Dhandare BC, Rather MA, Bhosale BP, Pawar R, Guttula PK, Pagarkar AU. Molecular modeling, docking and dynamic simulations of growth hormone receptor (GHR) of Labeo rohita. J Biomol Struct Dyn. 2022;40(7):3024-3037.
[CrossRef] [GoogleScholar] [PubMed]
- Barde RD. Bacterial diversity associated with Labeo rohita isolated from cultured and natural water bodies. Eur J Mol Clin Med. 2021;7(11):2020.
Citation: Mazhar MW, Raza A, Erdemli Kose SB, Iftikhar H, Tahir H, Zhou H, et al. (2022) Molecular and Morphological Identification of Fish gut Microbiota at Different Growth Stages of Labeo rohita. Clin Med Bio Chem. 8:119.
Copyright: © 2022 Mazhar MW et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.