Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 15, Issue 3
MecA and Panton Valentine Leukocidin (PVL) Gene Detection in Methicillin Resistant Staphylococcus aureus (MRSA) and Methicillin Resistant S. epidermidis (MRSE) from Children of Welfare Homes and Suburban Community in Perak, Malaysia
Nur Amirah Binti Ramli*, C. T. Soon, D. B. Singh and G. VivekananthanReceived: 18-Jun-2020, Manuscript No. JBP-24-5002; Editor assigned: 23-Jun-2020, Pre QC No. JBP-24-5002 (PQ); Reviewed: 07-Jul-2020, QC No. JBP-24-5002; Revised: 15-Jul-2024, Manuscript No. JBP-24-5002 (R); Published: 12-Aug-2024, DOI: 10.35248/2155-9597.24.15.515
Abstract
Staphylococcus aureus and S. epidermidis are the most commonly isolated bacteria on human skin that also colonize the upper respiratory tract of humans. They are gram-positive and appear as grape-like clusters under microscope. Both of S. aureus and S. epidermidis are normal flora of the human skin. They usually colonize and reside in or on part of human’s body without causing any infection. However, it can be pathogenic and cause serious infection when present in large numbers. They can lead to Skin and Soft-Tissue Infection (SSTIs). Previous studies have shown S. aureus to be the leading cause of bloodstream infections.
Keywords
Methicillin-Resistant Staphylococcus aureus (MRSA); Methicillin-Resistant S. epidermidis (MRSE); MecA gene; Panton-Valentine Leukocidin (PVL) gene; Penicillin-Binding Protein (PBPs); Children of welfare homes and suburban population
Introduction
Methicillin-Resistant Staphylococcus aureus (MRSA) causes potential infections and tend to become multidrug-resistant organism to β-lactam drugs [1]. According to Pantosti and Venditti it could be resistant towards other classes of antibiotics including lincosamides, aminoglycosides, macrolides, fluoroquinolones and tetracycline. Commonly, Penicillin- Binding Protein (PBPs) is found in normal bacteria that had an ability to bind with penicillin. However, MRSA differ from normal S. aureus in the presence of altered PBPs which is known as Penicillin- Binding Protein 2 (PBP2) or Penicillin-Binding Protein A (PBP2a) which prevents the binding of penicillin [2]. MRSA has spread rapidly in the health care environment until today. Abscesses, sore and boil on the skin will be the early symptoms of the mild-infection. Lack of proper sanitation and through contact with infected persons or objects can contribute to the infection.
The first case of MRSA was discovered in 1961, during the introduction of methicillin in the United Kingdom. Isolates were resistant towards all types of β-lactam antibiotics. In common bacterial cell, Penicillin-Binding Protein (PBPs) is a part of structure that serves to be the binding-site attachment for penicillin. However, production of foreign PBPs which is known as PBP2a or PBP2’ in MRSA, lowers the affinity towards methicillin and other β-lactam antibiotics. The presence of PBP2a is regulated by the expression of mecA gene. Two genes mecR1 and mecI control the expression of PBP2a within the mec region located upstream of the mecA gene. Hospital-Acquired MRSA (HA-MRSA) largely infects individuals with one or more comorbid condition or elderly patients. This will lead to possibilities of the patient developing pneumonia (infection of the lung passage), bacteremia (infection in bloodstream) or endocarditis (infection on the lining wall of the heart and valves). Surprisingly, Community-Acquired MRSA (CA-MRSA) has disseminated rapidly since 2000s in adults and children in various region and countries.
Materials and Methods
The university medical research ethics committee has approved this research (RCMP/MREC/2014/036) as attached in appendices section. The samples were collected from welfare homes and suburban populations in Kinta district, Perak [3].
In this cross sectional study, convenience sampling method was used. Convenience sampling method relies on data collection from population members who are conveniently available to participate in study. This method was selected as the logistics to do probability sampling was difficult and the yield would have been poor. The inclusion and exclusion criteria for the sampling are: i) individuals who are willing to take part in this research study, ii) individuals in a good health condition without having any chronic disease or receiving any special treatment. Exclusion criteria were: i) individual who were admitted into a health-care setting within the last 6 months and ii) individuals who are currently on medications.
All the participants who are involved were required to fill up the consent form before taking the nasal swab. Subjects gave their signature with date on the consent personally. While in welfare homes, consent was obtained from their caretakers. Demographic data of the subject such as their name, age, race and address were recorded for future use. Brief explanation on the swab procedure was given and consent was obtained from participants and guardians [4]. Guardians of the children from welfare homes were also included as they might serve as a reservoir for MRSA infections. In this process, sterile nasal swab, sterile distilled water, gloves, 70% alcohol and particular form details was prepared. Before the samples were taken, all the participants were briefed on the nasal swab procedure.
During swab procedure, the pointed sterile nasal swab was not touched on any surface to avoid environmental contamination. The cotton swab was gently inserted approximately 2 cm into the nares. Rotation in a circular motion against the anterior nasal mucosa for about 3 seconds was performed. Using the same swab, the same step was repeated for other nares. The swab was put back into the transport tube. It was ensured that the end of swab was pushed firmly and the cap secured. Tip of the swab need to be in contact with the moistened nutrient in the collection tube. Swab was collected from both of anterior nares as S. aureus commonly thrive in nasal cavities of humans. S. aureus was highly associated with Skin and Soft Tissue Infections (SSTIs). Colonization of S. aureus most highly indicates higher risk of subsequent infection including MRSA. All samples collected were labelled with number, name and age as in the consent form [5]. The samples collected were transported in an ice box to maintain the temperature at around 4°C and transported to the laboratory and stored in at 4°C.
Screening of S. aureus and S. epidermidis
For the detection of Staphylococcus aureus and S. epidermidis, Mannitol Salt Agar (MSA) was used as a selective media to inhibit bacteria other than S. aureus and S. epidermidis. Acid produced by mannitol-fermenting staphylococci changed the colour of the agar into yellow colonies suggesting S. aureus. Small pink colonies indicated S. epidermidis. For the identification of S. epidermidis, small growth of pinkish colony was observed. Gramstain smears of the organisms showing grape-like clusters that appeared as purple-coloured was presumed as S. epidermidis. In order to get the pure samples, the isolate was sub-cultured on NA which result as small white colonies after the 24 hours’ incubation at 37°C.
Phenotypic identification of S. aureus was confirmed by bactistaph latex agglutination kit test. It is a rapid test utilizing protein-coated latex particles which are capable of detecting both clumping factor and protein A simultaneously. It is recommended to distinguished S. aureus from other species of staphylococci. Aggregation of the smooth black latex suspension with the subsequent loss of black background visible to an unaided eye within 60 seconds, indicate positive result. Positive and negative control was included in the test [6].
Antibiotic Susceptibility Test (AST)
Mueller Hinton-Agar (MHA) was used in AST in accordance to clinical and laboratory standards institute guidelines. In this test, oxacillin (5 mg) and cefoxitin (30 mg) disk diffusion were used to test the susceptibility of isolates towards antibiotic resistant. Tryptic Soy Broth (TSB) was used to culture the isolates after 6-8 hours of incubation at 37°C. MacFarland standard 0.5 was used to compare and adjust the turbidity of bacterial suspension to get the standard bacterial number for microbial testing. This was done to avoid the suspension from being too heavy or too dilute which could lead to erroneous results [7].
100 μl of the culture was pipetted out and spread evenly in 3 different directions on Mueller-Hinton Agar (MHA) by using a sterile cotton swab. Antibiotic discs were gently placed on the MHA plate using a sterile syringe. Bunsen burner was used throughout the test to avoid any contamination. ATCC 33591 methicillin-resistant Staphylococcus aureus and ATCC 33862 methicillin-sensitive Staphylococcus aureus were used as positive and negative controls [8].
Phenotypic identification
Once an isolate was found resistant to cefoxitin or penicillin, it was tested for Penicillin-Binding Protein (PBP2) presence. The isolate was identified for the phenotype of methicillin-resistant Staphylococcus aureus and S. epidermidis. Instruction was followed as given in the manual. Isolates were identified as positive MRSA if there was an occurrence of agglutination in test latex within 3 minutes. On the other hand, isolates that produced no agglutination within 3 minutes, neither in test latex nor control latex was recognized as negative MRSA.
Detection of mecA gene
Detection of methicillin-resistant mecA gene was performed by following method of Siripornmongcolchai et al. The mecA gene was amplified with forward primer 5’- AAAATCGATGGTAAAGGTTGGC-3’ and reverse primer 5’- AGTTCTGCAGTACCGGATTTTGC-3’; the product base pair length is 286. Thermal cycling conducted as follows: 94°C-30 seconds; 58°C-30 seconds; 72°C-30 seconds for 30 amplification cycles.
Detection of 16S rRNA gene of S. epidermidis gene
Detection of 16S rRNA gene in S. epidermidis was carried out by using method describe by Frebourg et. al. The forward primer 5’- ATC AAA AAG TTG GCG AAC CTT TTC A-3’ and reverse primer 5’-CAA AAG AGC GTG GAG AAA AGT ATC A-3’ was used. An initial denaturation at 94°C for 2 min, followed by 30 cycles of amplification (denaturation at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 2 min) and ending with a final extension at 72°C for 5 min was set up [9].
Detection of PVL gene
Detection of PVL gene was performed by following method of Lina et. al. The PVL gene was amplified with primer sequence of forward primer luk-PV1, 5’- ATCATTAGGTAAAATGTCTGGACATGATCCA-3’ and reverse primer luk-PV-2 is 5’- GCATCAAATGTATTGGATAGCAAAAGC-3’; 433 base pair. Thermal cycling conducted was 30 seconds of denaturation at 94°C, annealing at 55°C; 1 min of extension at 72°C for 30 amplification cycles. PCR recipe was used as table shown below (Table 1).
| Populations | Samples (n) | Staphylococcus aureus | Staphylococcus epidermidis | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Suburban | 446 | M | 207 (46.4%) | 241 (54%) | M | 103 (42.7%) | 121 | M | 52 (43%) |
| F | 265 (59.4%) | F | 138 (57.3%) | F | 69 (57%) | ||||
| Children homes | 226 | M | 123 (54.4%) | 197 (87.2%) | M | 107 (54.3%) | 74 | M | 38 (51.4%) |
| F | 103 (45.6%) | F | 90 (45.7%) | F | 36 (48.6%) | ||||
| Total | 672 | 438 | 195 | ||||||
Table 1: S. aureus and S. epidermidis isolates from suburban and welfare homes.
Results and Discussion
A total of 672 samples were collected of which 446 samples were collected from suburban population and 226 samples from welfare homes. The table below represent sample collection of S. aureus and S. epidermidis based on gender for each population.
This study was conducted from suburban and welfare homes in Kinta district Ipoh, Perak, Malaysia. All samples were obtained from suburban populations (n=446) and children welfare homes (n=226). The gender distribution observed was 46.4% (207 individuals) were male and 59.4% (265 individuals) were female in the suburban population. While in welfare homes showed 54.4% (123 individuals) and 45.6% (103) were male and female respectively [10].
The results showed that 54% of S. aureus (241/446) and 27.1%(121/446) of S. epidermidis were isolated from suburban population. Welfare homes recorded 87.2% (197/226) of S. aureus and 41.6% (74/226) of S. epidermidis. The percentages of S. aureus carriers from welfare homes were higher by 33.2% as compared to suburban population. However, the data shows that isolations of S. epidermidis in welfare homes were 14.5% higher from suburban population.
The colonization of S. aureus in both populations (suburban: 54% and welfare homes: 87.2%) is considerably high as a study reported that S. aureus carriage among healthy individuals in Malaysia is 23.4%. This number also exceeds rates reported from several countries such as Lebanon (38.4%), USA (31.6%), Italy (25.9%) and Saudi Arabia (20.2%).
According to few studies, nasal carriage of S. aureus was higher among children in Italy (35%) aged 3-11 years, in Georgia (95%) aged 6-11 years and in Turkey (28.4%) aged 4-6 years.
Antibiotic Susceptibility Test (AST) results from suburban community and welfare homes were distinct. At early stage of this study, oxacillin was used as a preliminary assessment to screen the presence of mecA gene. However according to Felten et al., oxacillin required pure homogeneous MRSA/MRSE isolates. While according to Broekema et al., the occurrence of frequent hazy zones always misinterprets the susceptibility test as Methicillin-Susceptible S. aureus (MSSA). Low sensitivity and specificity of oxacillin was reported by Palazzo and Darini. Therefore, cefoxitin was used to detect isolates with mecAmediated resistance mechanism. Usage of cefoxitin is easier to interpret and more sensitive. Previous study also verified cefoxitin as more potent and correlate with the presence of mecA than when using oxacillin disc test.
In this finding, all the isolates from suburban community were susceptible to cefoxitin, ciprofloxacin, clindamycin, erythromycin, fosfomycin, fusidic acid, gentamicin, linezolid, oxacillin, quinipristin/dalfopristin, rifampicin, tigecycline, trimethoprim-sulphamethaxazole and vancomycin. Only 3 isolates (1.24%) of S. aureus and 8 isolates (6.6%) of S. epidermidis isolates were resistant to tetracycline, and only one (0.83%) of the S. epidermidis isolates was resistant towards chloramphenicol [11].
Whereas from welfare homes, one isolate (0.51%) of S. aureus isolates was resistant against cefoxitin, ciprofloxacin and oxacillin. Two (2/197) isolates (1.0%) were resistant against chloramphenicol and tetracycline. Three (3/197) isolates (1.52%) were resistant against gentamycin and four 4/197) isolates (2.0%) were resistant against erythromycin and linezolid. Five (5/197) isolates (2.54%) were resistant against fosfomycin, trimethoprim-sulphamethaxazole and vancomycin. Ten (10/197) isolates (1.97%) were resistant against rifampicin.
While for S. epidermidis, one isolate (1.4%) was resistant against linezolid, oxacillin and vancomycin. 2.7% (2/74) were resistant against gentamycin and 4.1% (3/74) isolates were resistant against cefoxitin, chloramphenicol and ciprofloxacin. Whereas 6.8% (5/74) were resistant against rifampicin and 8.1% (6/74) isolates were resistant against erythromycin and fosfomycin. Through the observation, resistance among both S. aureus and S. epidermidis was noticeably higher in welfare homes.
Subsequently, phenotypic identification of cefoxitin-resistant and oxacillin-resistant isolates were tested by using PBP2B kit. This technique was extensively shortened the MRSA identification time.
MecA gene is the gold standard used to detect MRSA. Phenotypic method such as disc diffusion and PBP2a test are widely used in microbiological protocol. However, they can be influenced by culture condition (temperature, medium pH and concentration of NaCl in medium) which may affect the results.
Initially, set primer of MR3 and MR4 producing 533 bp DNA fragment was used according to Siripornmongcolchai et al. Unfortunately, none of the isolates detected any targeted gene including the positive control of MRSA (ATCC 3351) and MRSE (ATCC 51625). Later, the primers were replaced with primer mA1 and mA2 as described by Katayama and Hiramatsu which targeted a part of mecA gene producing 286 bp DNA fragment. Four isolates of cefoxitin-resistant were successfully detected for the presence of mecA gene as shown in Figure 1. This indicates that the primer set mA1 and mA2 is more accurate and sensitive for PCR.

Figure 1: Percentage of antibiotic resistance of S. aureus isolates in both population.
In this study, only one isolate was resistant to oxacillin while the other three isolates were all resistant to cefoxitin. This showed that all four MRSA isolates had potential properties of CAMRSA which has low-level resistance to oxacillin and limited resistance against all non-β lactam antibiotics involved.
In order to endorse the three isolates (MRSE 3, 16 and 33) were strain of S. epidermidis, PCR was carried out as described by Frebourg, et al. These three isolates harboured the target 16S rRNA gene as shown in Figure 2.
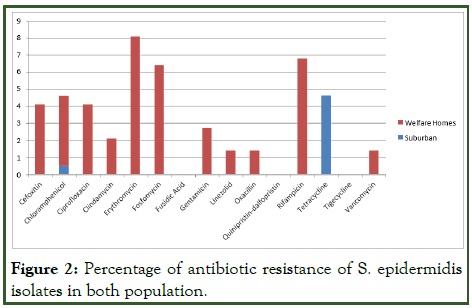
Figure 2: Percentage of antibiotic resistance of S. epidermidis isolates in both population.
Panton-Valentine Leukocidin (PVL) gene is considered as a one of the toxin genes associated with skin sepsis and necrotizing pneumonia. Its presence responsible for destruction of white blood cell. Widely reported CA-MRSA found to express this gene. Extensive cases reported that PVL gene is used a marker for CA-MRSA. PVL gene is believed to be more virulent than MSSA.
However, all the isolates were PVL negative as shown in Figure 3. Prior to the samples collection, it has been noticed that all the children involved did not have any skin infections. Our study correlated well with this fact and substantiated that PVL gene is mainly associated with primary skin or soft tissue infections, such as furuncles, abscesses or cellulitis (Figures 4 and 5).
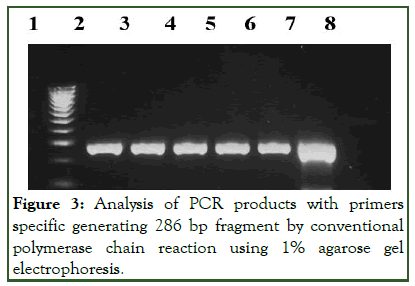
Figure 3: Analysis of PCR products with primers specific generating 286 bp fragment by conventional polymerase chain reaction using 1% agarose gel electrophoresis.
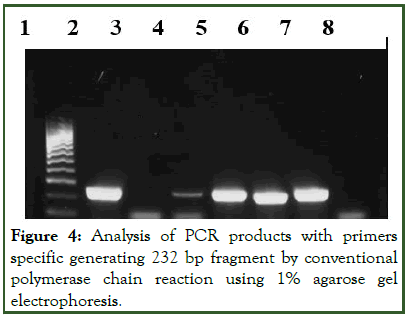
Figure 4: Analysis of PCR products with primers specific generating 232 bp fragment by conventional polymerase chain reaction using 1% agarose gel electrophoresis.
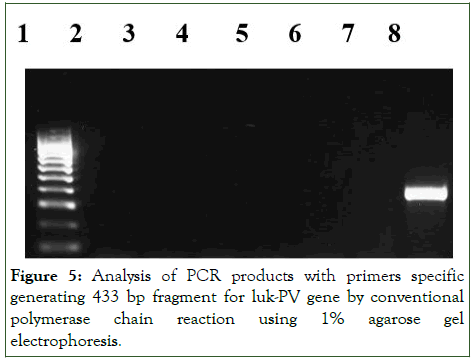
Figure 5: Analysis of PCR products with primers specific generating 433 bp fragment for luk-PV gene by conventional polymerase chain reaction using 1% agarose gel electrophoresis.
Nevertheless, there was study postulate that PVL gene is a poor marker for CA-MRSA. In fact, Neela, et al., study reported that only 4.5% nasal carriage of S. aureus (MSSA) harboured PVL gene. Recent report described by Bhatta, et al., that antibiotic resistance of PVL negative MRSA was higher as compared to PVL positive MRSA isolates. It was reported that PVL positive of MSSA strains contributed in large numbers to virulence factor as compared to MRSA.
According to Naimi, et al., CA-MRSA infections has low prevalence (<1%) in the United States among children and their guardians in New York city. Similarly, the occurrence of CAMRSA was also considerably low in Portugal (0.8%) among healthy young children aged <7 years. In addition, two isolates of one MRSA and one MRSE in the present study were from children aged 10 and 12 years. The occurrence of these infections may be due to their congested space, sharing facilities and personal belonging (eg: Toilet, bedroom, towel, toothbrush etc).
Two positive isolates of MRSA isolates were from the guardian of the children aged 42 and 61 years. This indicates that both children’s and guardian might have cross contamination due to close contact. The guardian might have had person to person contact with the children during the daily routine.
Previous studies conducted have shown that MRSA has been detected in intravenous drug abusers, people belonging to a lower socio-economic status and aboriginal groups. Overcrowded facilities, close contact and lack of proper sanitation have contributed to the spread of MRSA. Colonization of MRSA in individuals was associated with a 4- fold increase in the risk of infection.
The present study revealed that all the positive MRSA and MRSE isolates were from individuals of Indian ethnicity. This showed an agreement with the study reported by Ghaznavi-rad, et al., which stated the prevalence of MRSA was found to be significantly higher in the patients of Indian ethnicity [12].
CA-MRSA is more likely to be susceptible to non β-lactam antibiotic. Some researchers have proposed the increase in cases of CA-MRSA may be due to addition of mobile Staphylococcal Cassette Chromosome mec (SCCmec) type IV into PVL-positive strains. This finding indicates that nasal carriage of S. aureus in this investigation was high in healthy individual from suburban populations (54%) and welfare homes (87.2%). However, the resistance against all antibiotics used remains low and is considerably safe. It can be concluded that the prevalence of current CA-MRSA is sparse in comparison with previous findings derived in Malaysia. In addition, the present study corroborated that MRSA/MRSE prevalence (1.6%) is very low in the community or outside the healthcare setting. Nevertheless, their potential in antibiotic resistance and other virulent factors is a concern. Continued surveillance is still required.
Conclusion
In this study, convenient sampling method was used. However, the limitation of current study was not all children’s guardians were willing to participate in the sampling procedure. Thus, we only managed to collect the children’s nasal swab. In regard of laboratory procedures, conventional PCR was used and took up a lot of time instead of using real time PCR. Furthermore, other toxic genes that play an important.
References
- Brown DF, Walpole E. Evaluation of the mastalex latex agglutination test for methicillin resistance in Staphylococcus aureus grown on different screening media. J Antimicrob Chemother. 2001;47(2):187-189.
[Crossref] [Google Scholar] [PubMed]
- Brown DF, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, et al. Joint working party of the British society for antimicrobial chemotherapy, infection control nurses association. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother. 2005;56(6):1000-1018.
[Crossref] [Google Scholar] [PubMed]
- Chambers HF. The changing epidemiology of Staphylococcus aureus. Emerg Infect Dis. 2001;7(2):178-182.
[Crossref] [Google Scholar] [PubMed]
- Choi CS, Yin CS, Bakar AA, Sakewi Z, Naing NN, Jamal F, et al. Nasal carriage of Staphylococcus aureus among healthy adults. J Microbiol Immunol Infect. 2006;39(6):458-464.
[Google Scholar] [PubMed]
- David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616-687.
[Crossref] [Google Scholar] [PubMed]
- Frebourg NB, Lefebvre S, Baert S, Lemeland JF. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J Clin Microbiol. 2000;38(2):877-880.
[Crossref] [Google Scholar] [PubMed]
- Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M, et al. Association between Staphylococcus aureus strains carrying gene for panton-valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359(9308):753-759.
[Crossref] [Google Scholar] [PubMed]
- Graham PL, Lin SX, Larson EL. A US population-based survey of Staphylococcus aureus colonization. Ann Intern Med. 2006;144(5):318-325.
[Crossref] [Google Scholar] [PubMed]
- Halablab MA, Hijazi SM, Fawzi MA, Araj GF. Staphylococcus aureus nasal carriage rate and associated risk factors in individuals in the community. Epidemiol Infect. 2010;138(5):702-706.
[Crossref] [Google Scholar] [PubMed]
- Herman RA, Kee VR, Moores KG, Ross MB. Etiology and treatment of community-associated methicillin-resistant Staphylococcus aureus. Am J Health Syst Pharm. 2008;65(3):219-225.
[Crossref] [Google Scholar] [PubMed]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44(6):1549-1555.
[Crossref] [Google Scholar] [PubMed]
- Kateete DP, Kimani CN, Katabazi FA, Okeng A, Okee MS, Nanteza A, et al. Identification of Staphylococcus aureus: DNase and mannitol salt agar improve the efficiency of the tube coagulase test. Ann Clin Microbiol Antimicrob. 2010;9:1-7.
[Crossref] [Google Scholar] [PubMed]
Citation: Ramli NAB, Soon CT, Singh DB, Vivekananthan G (2024) MecA and Panton Valentine Leukocidin (PVL) Gene Detection In Methicillin Resistant Staphylococcus aureus (MRSA) and Methicillin Resistant S. epidermidis (MRSE) From Children of Welfare Homes and Suburban Community in Perak, Malaysia. J Bacteriol Parasitol. 15:515.
Copyright: © 2024 Ramli NAB, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

