Indexed In
- Open J Gate
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer
Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2023) Volume 12, Issue 3
Lastest Update on Therapeutics Strategies for the New Emerging Covid-19 variants. A Review
Liliana Elena Weimer*, Cattari G, Binelli A, Fanales Belasio E, Piras S and Sensi FReceived: 01-Dec-2022, Manuscript No. CMO-22-19075; Editor assigned: 05-Dec-2022, Pre QC No. CMO-22-19075 (PQ); Reviewed: 19-Dec-2022, QC No. CMO-22-19075; Revised: 22-Feb-2023, Manuscript No. CMO-22-19075 (R); Published: 01-Mar-2023, DOI: 10.35248/2327-5073.23.12.333
Abstract
The Coronavirus disease 2019 (COVID-19) pandemic has challenged public health around the world. Currently, there is an urgent need to explore antiviral therapeutic targets and effective clinical drugs. Monoclonal antibodies are a type of therapeutic agent under investigation for the treatment of COVID-19. These agents are often created by identifying pathogen-specific B cells of patients who have recently recovered from an infection or by immunizing mice genetically modified to have a humanized immune system and harvesting effective antibodies from them. Once the B cells are identified, the genes of immune globulin heavy and light chains are recovered. These genes are then expressed to produce monoclonal antibodies. Monoclonal antibodies have singular activity against a predetermined target; they therefore differ from convalescent plasma, which consists of polyclonal antibodies in serum derived from patients who are convalescing from an infection.
We summarized the latest therapeutic strategies against COVID-19, namely drugs targeting the SARS-CoV-2 life cycle and SARS-CoV-2 induced inflammation in host cells. The development of above two strategies is implemented by repurposing drugs and exploring potential targets.
A comprehensive summary of promising drugs, provides recommendations for clinicians as evidence-based medicine in the actual clinical COVID-19 treatment. Considering the emerging SARS-CoV-2 variants greatly impact the effectiveness of drugs and vaccines, we reviewed the appearance and details of SARS-CoV-2 variants at November 2022 for further perspectives in drug design, which brings updating clues to develop therapeutical agents against the variants. Based on this, the development of broadly antiviral drugs, combined with immune modulatory is prior to being considered for therapeutic interventions on mutant strains of SARS-CoV-2. Therefore, it is highly acclaimed the requirements of the concerted efforts from multi-disciplinary basic studies and clinical trials, which improves the accurate treatment of COVID-19 and optimizes the contingency measures to emerging SARS-CoV-2 variants.
Keywords
New drug target; SARS-CoV-2; Emerging COVID-19 variants; Monoclonal antibodies; Adaptive immunity
Abbreviations
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; COVID-19: Coronavirus Disease 2019; ACE2: Angiotensin Converting Enzyme 2; RBD: Receptor Binding Domain; CTD: C-Terminal Domain; NTD: N-Terminal Domain; RdRp: RNA Dependent RNA Polymerase; ERGIC: Endoplasmic Reticulum Golgi Intermediate Compartment; ARDS: Acute Respiratory Distress Syndrome; CRS: Cytokine Release Syndrome; MODS: Multiple Organ Dysfunction Syndrome; MAB: Monoclonal Antibody; Nbs: Nanobodies; JAK: Janus Kinase; STAT: Signal Transducer and Activator of Transcription; TCZ: Tocilizumab; α1AT: Alpha-1 Antitrypsin; CEP: Cepharanthine; TCM: Traditional Chinese Medicine; EMA: European Medicines Agency; FDA: U.S. Food and Drug Administration; NMPA: National Medical Products Administration; VOI: Variants of Interest; VOC: Variants of Concern; VUM: Variants under Monitoring.
Introduction
The new coronavirus was first identified on December 31, 2019. Due to being highly homologous to Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), 2019-nCoV was listed as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by the International Commission for Classification of Viruses (ICTV) on February 11, 2020. Among seven coronaviruses infecting humans, three coronaviruses can cause serious diseases, including Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) emerging in 2003, Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012, and Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in 2019 [1,2].
Vaccination represents the key strategy to control the COVID-19 pandemic through induction of neutralizing antibody responses and T cell-associated immunity that substantially decrease the risk of developing severe disease [3,4]. However, individuals who are immunocompromised (e.g. because of comorbidities, high age, or immunosuppressive treatment) might not mount a full adaptive immune response and thus remain susceptible. For individuals at high risk, individual Monoclonal Antibodies (mAbs) or cocktails of mAbs are administered as prophylaxis or therapy [5].
Literature Review
Monoclonal antibodies for SARS-CoV-2 infection
Monoclonal antibodies are a type of therapeutic agent under investigation for the treatment of COVID-19. These agents are often created by identifying pathogen-specific B cells of patients who have recently recovered from an infection or by immunizing mice genetically modified to have a humanized immune system and harvesting effective antibodies from them. Once the B cells are identified, the genes of immune globulin heavy and light chains are recovered. These genes are then expressed to produce monoclonal antibodies.
Monoclonal antibodies have singular activity against a predetermined target; they therefore differ from convalescent plasma, which consists of polyclonal antibodies in serum derived from patients who are convalescing from an infection). Monoclonal antibodies have been developed for the treatment and prophylaxis of other viral infections, such as HIV, influenza, RSV, MERS-CoV, Ebola and Zika virus. Of these, only monoclonal antibodies targeting RSV and Ebola have been shown to be effective in human trials (with the former having FDA approval). Several products targeting the other aforementioned viruses are currently being studied in clinical trials (Table 1).
| Approved drugs | ||||||||
|---|---|---|---|---|---|---|---|---|
| Product | Developer | Therapeutic class/drug type | Status | Approving authority | ||||
| Bebtelovimab | Ely-Llly | Monoclonal antibody | Authorization 12/2/2022 | FDA | ||||
| Kineret (Anakinra) | Sobi | Immunomodulator | Marketing authorization granted: 17/12/2021 | EMA | ||||
| Regkirona (Regdanvimab) | Celltrion healthcare | Monoclonal antibody | Marketing authorization granted: 12/11/2021 | EMA | ||||
| RoActemra | Roche | Immunomodulator | Marketing authorization for COVID-19 indication granted: 07/12/2021 | EMA | ||||
| Ronapreve (Casirivimab/Imdevimab) | Roche and Regeneron | Monoclonal antibody | Marketing authorization granted: 12/11/2021 | EMA | ||||
| Veklury (Remdesivir) | Gilead sciences | Nucleotide analogs | Conditional marketing authorization granted: 03/07/2020 | EMA | ||||
| Marketing authorization granted: 22/10/2020 | FDA | |||||||
| Xevudy (Sotrovimab) | GlaxoSmithKline and Vir Biotechnology | Monoclonal antibody | Marketing authorization granted: 17/12/2021 | EMA | ||||
| Developed drugs | ||||||||
| Product | Developer | Therapeutic class/drug type | Status | Data source | ||||
| Sarilumab | Sanofi Aventis | Immunomodulator | Clinical phase | EMA | ||||
| Canakinumab | Novartis | Monoclonal antibody | Clinical phase | Clinical trials | ||||
| Anakinra | Swedish Orphan Biovitrum AB (SOBI) | Immunomodulator | Clinical phase | EMA | ||||
| Tocilizumab | Roche | Immunomodulator | Clinical phase | EMA | ||||
| Pidotimod | / | Immunomodulator | / | / | ||||
| Cepharanthine | / | Immunomodulator | / | / | ||||
Table 1: Summary of approved and developed drugs against COVID-19.
The majority of direct antiviral monoclonal antibody products under development for SARS-CoV-2 target the spike protein, which the virus utilizes to enter host cells, thus blocking viral attachment and entry into human cells [6].
Products that have received FDA authorization include bamlanivimab/etesevimab, casirivimab/imdevimab (brand name REGEN-COV) and sotrovimab. However, because the omicron variant has become the dominant variant in the United States, bebtelovimab is the only monoclonal antibody currently recommended for use to treat COVID-19. Additionally, tixagevimab/cilgavimab (brand name Evusheld) is emergencyauthorized as pre-exposure prophylaxis against COVID-19 for immunocompromised individuals or those who cannot be vaccinated or mount post-vaccination immune response (Figure 1).
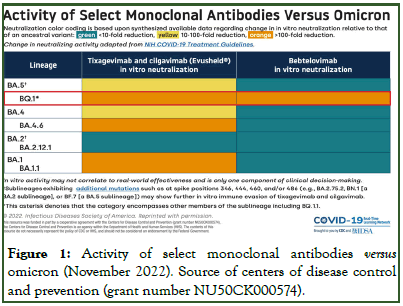
Figure 1: Activity of select monoclonal antibodies versus omicron (November 2022). Source of centers of disease control and prevention (grant number NU50CK000574).
The information is summarized from the available data online at sites and publication of advances in the development of therapeutic strategies against COVID-19 and perspectives in the drug design for emerging SARS-CoV-2 variants. Jialing Yin, Chengcheng Li, Chunhong Ye, Zhihui Ruan, Yicong Liang, Yongkui Li, Jianguo Wu, Zhen Luo, computational and structural biotechnology journal, volume 20,2022, pages 824-837.
Emerging omicron sub lineages BA.4.6, BA.2.75.2, BJ.1 (one parental lineage of the currently increasing XBB recombinant and all BQ.1.1 are probably resistant clinically used monoclonal antibodies
During the course of the COVID-19 pandemic, several SARS-CoV-2 lineages evolved mutations that confer partial or full resistance against some mAbs.
All monoclonal antibodies currently approved by the US Food and Drug Administration (FDA) or European Medicines Agency (EMA) target the Spike (S) protein. Consequently, only few mAbs remain suitable for treatment of individuals at high risk, and only bebtelovimab shows high efficacy against multiple omicron sublineages
However, novel omicron sublineages have been detected, harbouring additional S protein mutations within the epitopes of bebtelovimab and other mAbs. Novel sublineages include BA. 4.6 (with increasing incidence in several countries worldwide), BA.2.75.2 (with increasing incidence in India), BJ.1 (mainly observed in India and Bangladesh; notably BJ.1 is one parental lineage of the currently increasing XBB recombinant), and BQ. 1.1 (with increasing incidence in the USA and Europe) [6].
Recent investigations revealed that compared neutralization of omicron sub lineages BA.1, BA.4-5 (in which the amino acid sequence of the S protein is identical), BA.4.6, BA.2.75.2, BJ.1 and BQ.1.1 by single mAbs or mAb cocktails that are currently in clinical use, mAbs for which clinical use has been restricted or discontinued, and mAbs currently being evaluated in clinical trials. The study used pseudovirus particles (pp) that represent a suitable model to investigate SARS-CoV-2 cell entry and its neutralization [7].
As the investigators expected, pseudo virus particles bearing the BA.1 S protein (BA.1pp) were efficiently neutralised by bebtelovimab, adintrevimab, and cilgavimab-tixagevimab (50% Effective Concentration (EC50) <100 ng/ml), moderately neutralised by tixagevimab, romlusevimab, sotrovimab, and amubarvimab-romlusevimab (EC50 100 ng/ml-1000 ng/ml), and poorly neutralised by casirivimab, cilgavimab, amubarvimab, and casirivimab-imdevimab (EC50 1000 ng/ml-10000 ng/ml) [8]. Furthermore, BA4-5pp were efficiently neutralised by bebtelovimab and cilgavimab, moderately neutralised by imdevimab and cilgavimab-tixagevimab, and poorly neutralised by amubarvimab, romlusevimab, sotrovimab, casirivimab-imdevimab, and amubarvimab-romlusevimab, in line with expectations. For BA.4.6pp, bebtelovimab caused efficient neutralisation, whereas poor neutralisation was noted for imdevimab, amubarvimab, casirivimab-imdevimab, cilgavimab-tixagevimab, and amubarvimab-romlusevimab. With BA.2.75.2pp, bebtelovimab caused efficient neutralisation, whereas regdanvimab and sotrovimab caused poor neutralisation. For BJ.1pp, none of the tested mAbs or mAb cocktails caused high neutralization, whereas casirivimab, tixagevimab, sotrovimab, and cilgavimab-tixagevimab showed moderate neutralisation, and amubarvimab, casirivimabimdevimab, and amubarvimab-romlusevimab caused poor neutralization. Finally, none of the tested mAbs or mAb cocktails caused appreciable neutralization of BQ.1.1pp (Figure 2).
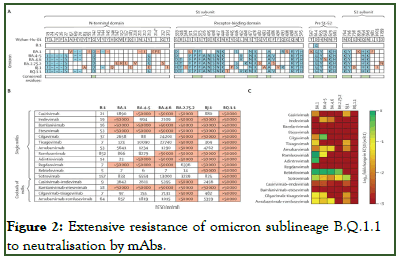
Figure 2: Extensive resistance of omicron sublineage B.Q.1.1 to neutralisation by mAbs.
The data base reveal that emerging omicron sublineages are resistant to most (i.e., BA.4.6, BA.2.75.2, and BJ.1) or all (BQ. 1.1) clinically used mAbs.
As a consequence, in patients at high risk, treatment with mAbs alone might not provide a therapeutic benefit in regions of the globe in which BQ.1.1 is spreading, suggesting that additional treatment options (e.g. paxlovid or molnupiravir) should be considered. Furthermore, novel, broadly active mAbs are urgently needed for prophylactic or therapeutic treatment, or both, in patients at high risk.
A) Location of mutations (blue and red) in the spike proteins of SARS-CoV-2 lineages B.1, BA.1, and BA.4–5 (which are identical at the amino acid level), BA.4.6, BA.2.75.2, BJ.1, and BQ.1.1 (numbered according to the spike protein of SARS-CoV-2 Wuhan-Hu-01). Mutations that are unique to only one of the omicron sublineages are highlighted in red and conserved mutations among omicron sublineages are indicated beneath the sequences in green;
B) Pseudo virus particles carrying the indicated S proteins were preincubated with different concentrations of single mAbs or cocktails of mAbs, before being inoculated onto vero cells. pseudo virus entry was analyzed at 16 h-18 h post-inoculation, by measuring firefly luciferase activity in cell lysates, and was normalized against samples without any antibodies (0% inhibition). The EC50 was calculated by use of a non-linear regression model. Data represent the mean of three biological replicates (performed with four technical replicates). For additional information see the appendix (p 12);
C) Heat map indicating the fold change in EC50 compared with B.1 pseudovirus particles. EC50=the concentration required for 50% of maximum inhibition. mAbs=monoclonal antibodies. Pre S1-S2=the domain between the receptor-binding domain and the S1-S2 cleavage site. S=spike. *The BA.1 spike protein contains a unique insertion at position 214 (EPE).
Early treatment and disease stage
The clinical course of COVID-19 is heterogeneous and may be prolonged. A major challenge with drug development has been tailoring therapies to the stage of disease. Most clinical trial data currently apply to hospitalized patients. There is limited insight into effective treatments for the early stages or in the post hospital or long COVID setting.
There is a window of missed opportunity: Early treatment, by reducing progression to hospitalization, might substantially reduce long-term morbidity and mortality and reduce the burden on healthcare resources. For an early-intervention strategy to work, we need to accurately identify patients at greatest risk of clinical deterioration. Demographic factors help, as well as laboratory measures. Deep phenotyping studies, incorporating both clinical features and immunological biomarkers and utilizing machine learning techniques, are likely to deliver a more sophisticated understanding necessary to identify best targets for early therapy.
Established therapies and future opportunities for intervention early in disease. Following infection with SARS-CoV-2, there are specific time points in the disease trajectory where different therapies could be optimally administered. At this time, the majority of treatments have been targeted during hospitalization and particularly at late stages of acute disease during ICU admission. Additionally, many of the therapies trialed are antibody therapies and are cost prohibitive, especially in low-to middle-income countries. There is currently a window of missed opportunity early in disease to reduce progression to hospitalization.
Long COVID
An important emerging issue is the symptom complex long- COVID. Prevalence appears high in both hospitalized patients and those with mild/moderate disease [9]. Evidence suggests that long-COVID is more severe than other post viral syndromes. Patients admitted to hospital with COVID-19 had significantly higher rates of death, cardiovascular disease, neurological and mental health disorders, fatigue, and coagulation disorders than those admitted with influenza [10].
The mechanism of long-COVID is not well understood. Theories include long-term cellular damage caused by the SARS-CoV-2 virus during acute infection, SARS-CoV-2 persistence, post-acute illness, and the development of autoimmunity [10,11]. Changes to social circumstances, including job loss and social isolation, may also negatively impact the clinical course. Without a clear understanding of the pathogenesis of long-COVID, the development of effective therapy is challenging.
Encouragingly, studies are shedding light on the pathogenesis of the most common complaints in long-COVID, namely pulmonary and neurologic symptoms. The highly sensitive hyperpolarized Xenon magnetic resonance imaging method of visualizing microstructure abnormalities in the lung shows persistent anomalies at the alveolar-endothelial-epithelial junction. Neuropathology studies, mainly from autopsies, show abnormalities similar to dementia, schizophrenia, and depression. Brain specimens show evidence of widespread inflammatory changes, including lymphoid infiltration and microglial activation [12]. Coagulation abnormalities are also common [13], with brain infarcts seen in about 20% of individuals and micro thrombi and hemorrhages seen in 15% of individuals. These abnormalities might help to explain the symptoms of “brain fog” and fatigue in those with long-COVID.
Therapeutics currently under investigation for long-COVID target different proposed mechanisms for pathogenesis. Strategies include anti fibrotic medication due to the observation of fibrotic lung changes, anticoagulants, statins for their anti-inflammatory properties, and dedicated long-COVID rehabilitation centers. Observational data also suggests improvement in long-COVID symptoms post-vaccination, the theory being that vaccination induces elimination of residual SARS-CoV-2 virus [14].
Vulnerable populations
Low to middle income countries will likely bear the greatest disease burden from COVID-19 due to delayed access to effective vaccination and the consequences of structural inequity, such as overcrowding. These countries need affordable, easily administered COVID-19 therapies, such as temperature stable oral medicines or inhaled therapeutics. Robust and accountable collective purchasing and distribution systems will need to stand independently of national interest. Collective drug purchasing, driven by the WHO and nongovernment initiatives, like the global fund and the gates foundation, has proven effective in ongoing challenges, such as HIV/AIDS. A similar approach in COVID-19 is necessary, accompanied by preventative strategies and culturally appropriate community engagement, particularly as COVID-19 will likely persist as a global threat until all countries can sustainably access treatment.
Even with widespread COVID-19 vaccination uptake, there will always be people with poor vaccination response. Immunomodulatory medications like glucocorticoids and rituximab lead to reduced response to SARS-CoV-2 vaccines, and it is possible that other immune-modifying therapies cause a similarly reduced response. Without herd immunity, these patients will remain vulnerable to severe infection. Huge resources have appropriately been allocated to the development and distribution of effective vaccines, and this remains one of the great successes of the pandemic. Despite this, effective and cheap therapeutics are critical given viral genome evolution, particularly for those who do not sufficiently respond to vaccines, for those who decline to receive vaccines, for those yet to be able to access vaccines on scale, and as a back-up should variants of concern evade vaccine-induced immunity. While vaccine development has been rightfully relatively prioritized to this point, the efforts of the medical and scientific community will only be complete with a renewed focus on therapeutics.
This focus on therapeutics will be particularly important for certain populations. Patients on B cell-depleting therapies, used widely as a treatment in hematological malignancy, rheumatology, clinical immunology, and neurology, are particularly vulnerable. B cell-depleting therapy has been identified as a significant inhibitor of COVID-19 vaccine serological responses, but our understanding of cell-mediated immunity is more primitive. For these patients, and for those with inherited or acquired immunodeficiency, preexposure prophylaxis with long-acting neutralizing monoclonal antibodies clearly has a role, given that time is critically of essence for antibody generation if one wants to survive infection. Given that this approach does not rectify the underlying inadequate host immune response, efforts should also be focused on how we can prime these abnormal immune systems in focal ways to generate some immunity. This will be both a scientific and a logistical challenge, given the relative few who stand to benefit, but success in these efforts will likely have huge ramifications for those who are born with immune deficiencies or inherit them from treating cancer and autoimmune disease, and the broader measures required to protect them.
The COVID-19 therapeutic landscape
At different stages of COVID-19, pathology and relevant targets differ. In the early stages, antivirals are key, compared to later in the disease where inflammation and its consequences are largely culpable. Regrettably, the important timing of therapy administration within relevant windows of opportunity remains inexact. In fact, while some overall success has been achieved in all therapeutic classes, the choice and best use of agents is evolving rapidly.
Results
Therapeutics strategies and monoclonal antibodies
Bebtelovimab: In February 2022, FDA issued an emergency use authorization for bebtelovimab for the treatment of mild to moderate COVID-19 in adults and certain pediatric patients over the age of 12 for whom alternative treatment options are not accessible or clinically appropriate.
Available in vitro data from both pseudotyped virus like particle and authentic SARS-CoV-2 virus neutralization testing shows that this monoclonal antibody retains neutralization efficacy against all variants of SARS-CoV-2 except Mu. It retains efficacy against the omicron variant of SARS-CoV-2, including BA. 1.1.529, BA.1.1 and BA.2, BA.4, and BA.5 (FDA fact sheet for health care providers, June 2022).
Clinical data for its EUA authorization came from the BLAZE-4 trial, which was performed prior to the era when omicron was a dominant strain (50% of participants had delta, and 29% had Alpha). BLAZE-4 was a phase 2 study that enrolled both low risk and high risk nonhospitalized participants with mild to moderate COVID-19 and examined clinical efficacy of 175 mg bebtelovimab alone and combined with bamlanivimab/etesevimab compared to placebo in preventing hospitalization or death, when administered within 3 days of testing positive.
The study design was complex, with many arms, and included a randomized portion and an open-label portion. One notable difference between the treatment groups was that more participants in the placebo arm were seropositive at baseline than in the group receiving bamlanivimab/etesevimab/bebtelovimab together or the group receiving bebtelovimab alone (15%, 9% and 7%, respectively); all participants were unvaccinated. A nonclinical, virologic primary endpoint of persistently high viral load by day 7 was chosen.
A persistently high viral load was observed in 21%, 13% and 14% of participants in the placebo, combination monoclonal antibody and bebtelovimab groups, respectively. The secondary endpoint of COVID-19-related hospitalization or death by day 29 was observed in 1.6%, 2.4% and 1.6% of those three groups, respectively.
Tixagevimab/cilgavimab: Tixagevimab/cilgavimab (brand name EVUSHELD; AZD7442) is a long-acting monoclonal antibody combination that has been FDA authorized for emergency use as pre-exposure prophylaxis for prevention of COVID-19 in certain immunocompromised adults and pediatric patients since December 2021 (dosing was revised in February 2022).
Although the combination was highly active against earlier SARS-CoV-2 variants, in October 2022 FDA announced that individuals receiving tixagevimab/cilgavimab face an increased risk for developing COVID-19 when exposed to novel variants of SARS-CoV-2 that evade neutralization because tixagevimab has no activity against circulating BA.4/BA.5 sub variants, while cilgavimab has reduced activity.
IDSA guidelines continue to suggest pre-exposure prophylaxis with tixagevimab/cilgavimab rather than no tixagevimab/cilgavimab in moderately or severely immunocompromised individuals at increased risk for inadequate immune response to COVID-19 vaccine or for whom COVID-19 vaccine is not recommended due to a documented serious adverse reaction to the vaccine (conditional recommendation, low certainty of evidence).
Based on current U.S. variant prevalence, NIH guidelines continue to recommend tixagevimab/cilgavimab as pre-exposure prophylaxis for patients who are moderately to severely immunocompromised and may have inadequate immune response to COVID-19 vaccination (BIIa) or are not able to be fully vaccinated with any available COVID-19 vaccines due to a documented history of severe adverse reaction to a COVID-19 vaccine or any of its components (AIIa).
Bamlanivimab/etesevimab: Eli Lilly’s monoclonal antibody bamlanivimab (also known as LY-CoV555, aka LY3819253) was originally derived from the blood of one of the first U.S. patients who recovered from COVID-19. It is a recombinant neutralizing monoclonal antibody directed against the SARS-CoV-2 spike protein. Eli Lilly’s etesevimab (LY-CoV016, aka JS016, aka LY3832479) is a monoclonal antibody directed against the SARS-CoV-2 surface spike protein’s receptor binding domain.
Casirivimab/imdevimab: Regeneron’s REGEN-COV (previously known as REGN-CoV2 or REGEN-CoV2) consists of two antibodies that bind to different regions of the SARS-CoV-2 spike protein receptor binding domain: Casirivimab (REGN10933) and imdevimab (REGN10987). In October 2020, an independent data monitoring committee recommended halting a study examining the use of casirivimab plus imdevimab in hospitalized patients requiring high-flow oxygen or mechanical ventilation, due to a potential safety signal and an unfavorable risk/benefit profile.
However, in outpatients, two double-blind, randomized, controlled trials of REGEN-COV (trials 2067 and 20145; also see weinreich, 2021, an interim analysis) have recently released results via press release. In study 20145 (N=803), an outpatient dose-ranging virologic efficacy trial of REGEN-COV among low-risk outpatients with asymptomatic or mild symptomatic COVID-19, there was comparable viral load drop among all dose levels, including the subcutaneously dosed groups, through day 7 (without a dose-response effect). In the large outpatient study of the impact of REGEN-COV on clinical outcomes among high-risk patients with COVID-19 (N=4,567), there was a significant reduction in COVID-related hospitalization or death of 71.3% (1.3% vs. 4.6%; p<0.0001) in the 2,400 mg group and 70.4% (1.0% vs. 3.2%) in the 1,200 mg group, as compared to placebo. The effect was strongest among those with baseline SARS-CoV-2 viral load above 1 million and negative SARS-CoV-2 antibodies. Likewise, the time to symptom resolution was faster with both doses than placebo, with a median of 10 versus 14 days to clinical improvement (p<0.0001). In a large safety database of participants from these two trials (N=6,334), no serious safety signal of concern was observed, and severe adverse events were less frequent in the combined monoclonal antibody group than in the placebo group (1.4% vs. 4%). The large, phase 3 U.K. NHS RECOVERY trial then evaluated regen-cov in a very large (N=9,785) group of patients hospitalized with COVID-19, and found that, among participants who were seronegative for SARS-CoV-2 at baseline, there was a significant mortality benefit from receiving the combination of casirivimab and imdevimab (REGEN-COV), as compared to standard of care.
REGEN-COV has also been studied as a “passive vaccine” in study 2069 to prevent infection among household contacts of patients with known COVID. Final phase 3 results are forthcoming and showed a reduction in symptomatic (SARS-CoV-2 PCR+) infections from 8/233 (3.6%) in placebo to 0/186 (0%) in REGEN-COV group (p<0.01). There was also a reduction in SARS-CoV-2 PCR+ infections of any symptom degree with a high (above 10,000) viral load, from 13/212 (6.1%) in the placebo group to 0/179 (0%) in the REGEN-COV group (p<0.001).
In November 2020, FDA granted emergency use authorization for both bamlanivimab and the combination of casirivimab and imdevimab in outpatients with mild to moderate COVID-19 who are at high risk for severe COVID-19. These approvals were based on interim analyses of outpatient randomized controlled trials, which showed a reduction in COVID-19 related hospitalization or emergency room visits with the use of these monoclonal products. Following these two trials, the randomized controlled ACTIV-3 study of bamlanivimab in hospitalized COVID-19 patients without end-organ failure showed little additional impact on sustained recovery over 90 days when compared to placebo plus standard of care, which included remdesivir and corticosteroids (Gottlieb, January 2021). The study was stopped by the data safety and monitoring board for futility after 314 participants had been enrolled.
Sotrovimab: In May 2021, FDA approved for emergency use a new monoclonal anti-SARS-CoV-2 antibody manufactured by GSK and Vir Biotechnology, sotrovimab (formerly VIR-7831), which binds to a highly conserved epitope of the receptor binding domain of viral spike protein. The approval was based on an analysis of data from the COMET-ICE study, a randomized controlled trial investigating the safety and efficacy of sotrovimab 500 mg IV given within 5 days of symptom onset in 583 nonhospitalized adults with mild to moderate SARS-CoV-2 infection. The primary endpoint, death or >24 hours of hospitalization through day 29 after dose, was reached in 21 of 292 (7%) patients in the placebo group compared to 3 of 291 (1%) who received sotrovimab, an 85% reduction (p=0.002). Based on these results at the time of the interim analysis, an independent safety and data monitoring committee recommended stopping the study. A parallel in-vitro study found that sotrovimab retains efficacy against most circulating variants. Further studies are planned by the manufacturers, including a pharmacokinetic study (comet-peak) comparing venous and intramuscular sotrovimab, which could lay the groundwork for intramuscular administrations of anti-SARS-CoV-2 monoclonal antibodies, rather than intravenous infusions; this will be followed by phase 3 trials assessing the impact of intramuscular sotrovimab on hospitalization and death in high-risk people with COVID-19 and on symptomatic infection among asymptomatically infected individuals (comet-tail and comet-star).
Emerging SARS-CoV-2 variants as of 30 November 2022: SARS-CoV-2 has been continuing to evolve, posing higher infectivity efficiency and faster transmission, leading to a greater risk to global public health. To better assess the consequences of different variants and facilitate prevention measures or medical countermeasures, WHO divides them into Variants Of Interest (VOI) and Variants Of Concern (VOC) [15]. There are currently two VOIs: Kappa and Mu; and five VOCs: Alpha, beta, gamma, delta, and omicron (Table 2). On November 24, 2021, a new SARS-CoV-2 variant B.1.1.529 named omicron was discovered in South Africa. Previously, the SARS-CoV-2 delta variant has become the main epidemic strain in many countries [16]. Now, the emergence of omicron has aroused new attention and vigilance. The mutations in omicron are concentrated in the S protein, and there seems to be a tendency to collect mutations that are beneficial to immune escape [17,18]. A model predicts and calculates that omicron’s infectivity is about ten times that of the original virus or twice that of the delta variant. Omicron may greatly undermine the efficacy of the Eli Lilly Monoclonal Antibody (mAb) approved by the FDA, and may also reduce the efficacy of mAbs from Celltrion and Rockefeller university [19]. The main SARS-CoV-2 variants reported in different places worldwide create new concerns about anti-viral drugs and vaccinations against COVID-19 pandemic. The following critical mutation sites in SARS-CoV-2 genome determine the virulence and spread of the SARS-CoV-2, which provides fresh ideas in the drug design for the main emerging variants [20].
Variants Of Concern (VOC)
For these variants, clear evidence is available indicating a significant impact on transmissibility, severity and/or immunity that is likely to have an impact on the epidemiological situation in the EU/EEA. The combined genomic, epidemiological, and in vitro evidence for these properties invokes at least moderate confidence. In addition, all the criteria for variants of interest and under monitoring outlined below apply (Table 2).
| WHO label | Lineage + additional mutations | Country first detected (community) | Spike mutations of interest | Year and month first detected | Impact on transmissibility | Impact on immunity | Impact on severity | Transmission in EU/EEA |
|---|---|---|---|---|---|---|---|---|
| Omicron | BA.2 | South Africa | (y) | Nov 21 | Increased (v) (1, 2) | Increased (v) (3) | Reduced (v) (4, 5) | Community |
| Omicron | BA.4 | South Africa | L452R, F486V, R493Q | Jan 22 | No evidence | Increased (6, 7) | No evidence | Community |
| Omicron | BA.5 | South Africa | L452R, F486V, R493Q | Feb 22 | No evidence | Increased (6, 7) | Unclear (8) | Dominant |
| y: G142D, N211I, Δ212, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K | ||||||||
| All sub-lineages of the listed lineages are also included in the variant, e.g., BA.1.1 is included in Omicron BA.1 as it is a sub-lineage of BA.1. | ||||||||
Table 2: Variants Of Concern (VOC).
Variants of Interest (VOI)
For these variants, evidence is available on genomic properties, epidemiological evidence or in vitro evidence that could imply a significant impact on transmissibility, severity and/or immunity, realistically having an impact on the epidemiological situation in the EU/EEA. However, the evidence is still preliminary or is associated with major uncertainty. In addition, all the criteria for variants under monitoring outlined below apply (Table 3).
| WHO label | Lineage + additional mutations | Country first detected (community) | Spike mutations of interest | Year and month first detected | Impact on transmissibility | Impact on immunity | Impact on severity | Transmission in EU/EEA |
|---|---|---|---|---|---|---|---|---|
| Omicron | BA.2.75 | India | (y) | May 22 | No evidence | Increased (9, 10) | No evidence | Detected (a) |
| Omicron | BQ.1 | n/a | K444T, N460K | n/a | No evidence | Increased (m) (11) | No evidence | Detected (a) |
| x: Any amino-acid substitution; y: W152R, F157L, I210V, G257S, D339H, G446S, N460K, Q493 (reversion) |
||||||||
Table 3: Variants of Interest (VOI).
Variants under monitoring
These additional variants of SARS-CoV-2 have been detected as signals through epidemic intelligence, rules-based genomic variant screening, or preliminary scientific evidence. There is some indication that they could have properties similar to those of a VOC, but the evidence is weak or has not yet been assessed by ECDC. Variants listed here must be present in at least one outbreak, detected in a community within the EU/EEA, or there must be evidence that there is community transmission of the variant elsewhere in the world (Table 4).
| WHO label | Lineage + additional mutations | Country first detected (community) | Spike mutations of interest | Year and month first detected | Impact on transmissibility | Impact on immunity | Impact on severity | Transmission in EU/EEA |
|---|---|---|---|---|---|---|---|---|
| Omicron | B.1.1.529+ R346 X (x) | n/a | R346 X | n/a | No evidence | No evidence | No evidence | Detected (a) |
| Omicron | B.1.1.529+ K444 X, N460 X | n/a | K444 X, N460 X | n/a | No evidence | Increased (m) (11) | No evidence | Detected (a) |
| Omicron | B.1.1.529+ N460 X, F490 X (y) | n/a | N460 X, F490 X | n/a | No evidence | Increased (m) (11) | No evidence | Detected (a) |
| x: Any amino-acid substitution; y: Including XBB and other sub-lineages |
||||||||
Table 4: Variants under monitoring.
De-escalated variants
These additional variants of SARS-CoV-2 have been de-escalated based on at least one the following criteria.
• The variant is no longer circulating;
• The variant has been circulating for a long time without any
impact on the overall epidemiological situation;
• Scientific evidence demonstrates that the variant is not
associated with any concerning properties (Table 5).
| WHO label | Lineage + additional mutations | Country first detected (community) | Spike mutations of interest | Year and month first detected | Impact on transmissibility | Impact on immunity | Impact on severity | Rationale for de-escalation |
|---|---|---|---|---|---|---|---|---|
| Alpha | B.1.1.7 | United Kingdom | N501Y, D614G, P681H | Sep 20 | Increased (v) (12) | Similar | Increased (v) (13, 14) | Drastically reduced circulation in the EU/EEA following the emergence of delta; little evidence of impact on vaccine induced immunity |
| n/a | B.1.1.7+E484K | United Kingdom | E484K, N501Y, D614G, P681H | Dec 20 | Increased (v) (12) | Increased (v) (15, 16) | Increased (v) (13) | Very low levels of circulation in the EU/EEA |
| Epsilon | B.1.427/B.1.429 | USA | L452R, D614G | Sep 20 | Unclear (17) | Increased (v) (17) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA and available data indicating that vaccines and treatments are effective against such variant |
| n/a | B.1.616(c) | France | V483A, D614G, H655Y, G669S | Feb 21 | Detection (c) (18) | No evidence | No evidence | Not detected since 2021-04-23 (19) |
| Eta | B.1.525 | Nigeria | E484K, D614G, Q677H | Dec 20 | No evidence | Increased (m) (15, 20) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| Theta | P.3 | The Philippines | E484K, N501Y, D614G, P681H | Jan 21 | Increased (m) (12) | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| Kappa | B.1.617.1 | India | L452R, E484Q, D614G, P681R | Dec 20 | Increased (v) (21) | Increased (v) (22-25) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.620 | Unclear (b) | S477N, E484K, D614G, P681H | Feb 21 | No evidence | Increased (m) (15, 26) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.617.3 | India | L452R, E484Q, D614G, P681R | Feb 21 | Increased (m) (12) | Increased (m) (15, 17) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.214.2 | Unclear2 | Q414K, N450K, ins214TDR, D614G | Dec 20 | No evidence | No evidence | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | A.23.1+E484K | United Kingdom | V367F, E484K, Q613H | Dec 20 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | A.27 | Unclear (b) | L452R, N501Y, A653V, H655Y | Dec 20 | Increased (m) (12) | Increased (m) (17) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | A.28 | Unclear (b) | E484K, N501T, H655Y | Dec 20 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | C.16 | Unclear (b) | L452R, D614G | Oct 20 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.351+P384L | South Africa | P384L, K417N, E484K, N501Y, D614G, A701V | Dec 20 | Increased (v) (27) | Increased (v) (28, 29) | Unclear (30) | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.351+E516Q | Unclear (b) | K417N, E484K, N501Y, E516Q, D614G, A701V | Jan 21 | Increased (v) (27) | Increased (v) (28, 29) | Unclear (30) | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.1.7+L452R | United Kingdom | L452R, N501Y, D614G, P681H | Jan 21 | Increased (v) (12) | Increased (m) (17) | Increased (v) (13) | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.1.7+S494P | United Kingdom | S494P, N501Y, D614G, P681H | Jan 21 | Increased (v) (12) | Increased (m) (31) | Increased (v) (13) | No longer detected or detected at extremely low levels in the EU/EEA |
| Iota | B.1.526 | USA | E484K, D614G, A701V | Dec 20 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.526.1 | USA | L452R, D614G | Oct 20 | No evidence | Increased (m) (17) | No evidence | Lineage withdrawn from pango |
| n/a | B.1.526.2 | USA | S477N, D614G | Dec 20 | No evidence | No evidence | No evidence | Lineage withdrawn from pango |
| Zeta | P.2 | Brazil | E484K, D614G | Jan 21 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.1.519 | Mexico | T478K, D614G | Nov 20 | No evidence | Increased (m) (17) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | AV.1 | United Kingdom | N439K, E484K, D614G, P681H | Mar 21 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | AT.1 | Russian Federation | E484K, D614G, N679K, ins679GIAL | Jan 21 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | C.36+L452R | Egypt | L452R, D614G, Q677H | Dec 20 | No evidence | Increased (m) (17) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | P.1+P681H | Italy | D614G, E484K, H655Y, K417T, N501Y, P681H | Feb 21 | No evidence | Unclear (32, 33) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| Mu | B.1.621 | Colombia | R346K, E484K, N501Y, D614G, P681H | Jan 21 | Increased (m) (12) | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| Lambda | C.37 | Peru | L452Q, F490S, D614G | Dec 20 | No evidence | Increased (v) (34, 35) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | AY.4.2 | United Kingdom | L452R, T478K, D614G, P681R, A222V, Y145H | Jun 21 | Increased (v) (36) | Similar (v) (36, 37) | Similar (v) (36) | Delta sub-lineages will continue to be monitored within delta VOC |
| n/a | B.1.1.318 | Unclear (b) | E484K, D614G, P681H | Jan 21 | No evidence | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.617.2 + K417N | United Kingdom | L452R, T478K, D614G, P681R, K417N | Jun 21 | No evidence | No evidence | No evidence | Delta sub-lineages will continue to be monitored within delta VOC |
| n/a | C.1.2 | South Africa | D614G, E484K, H655Y, N501Y, N679K, Y449H | Jun 21 | Increased (m) (12) | Increased (m) (15) | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.617.2 + E484X (d) | India | L452R, T478K, D614G, P681R, E484X (d) | Apr 21 | No evidence | No evidence | No evidence | Delta sub-lineages will continue to be monitored within delta VOC |
| n/a | B.1.617.2 + Q613H | India | L452R, T478K, D614G, P681R, Q613H | Apr 21 | No evidence | No evidence | No evidence | Delta sub-lineages will continue to be monitored within delta VOC |
| n/a | B.1.617.2 + Q677H | India | L452R, T478K, D614G, P681R, Q677H | Apr 21 | No evidence | No evidence | No evidence | Delta sub-lineages will continue to be monitored within delta VOC |
| Beta | B.1.351 | South Africa | K417N, E484K, N501Y, D614G, A701V | Sep 20 | Increased (v) (27) | Increased (v) (28, 29) | Increased (v) (14, 30) | No longer detected or detected at extremely low levels in the EU/EEA |
| Gamma | P.1 | Brazil | K417T, E484K, N501Y, D614G, H655Y | Dec 20 | Increased (v) (38) | Increased (v) (39) | Increased (v) (14) | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | B.1.640 | The Republic of Congo | D614G, F490R, N394S, N501Y, P681H, R346S, Y449N, 137−145de | Sep 21 | No evidence | No evidence | No evidence | No longer detected or detected at extremely low levels in the EU/EEA |
| n/a | XF | United Kingdom | Omicron-like | Jan 22 | No evidence | No evidence | No evidence | No longer detected. |
| n/a | XD | France | NTD delta-like; remaining omicron-like | Jan 22 | No evidence | No evidence | No evidence | No longer detected. |
| Delta | B.1.617.2 | India | L452R, T478K, D614G, P681R | Dec 20 | Increased (v) (40) | Increased (v) (41-43) | Increased (v) (42, 44) | Detected at extremely low levels in the EU/EEA |
| Omicron | BA.1 | South Africa and Botswana | (x) | Nov 21 | Increased (v) (1, 45) | Increased (v) (3,46,47) | Reduced (v) (48-50) | Detected at extremely low levels in the EU/EEA |
| Omicron | BA.3 | South Africa | (z) | Nov 21 | No evidence | No evidence | No evidence | Detected at extremely low levels in the EU/EEA |
| Omicron | BA.2 + L452X (x) | n/a | L452X | n/a | No evidence | Increased (7) | No evidence | Detected at extremely low levels in the EU/EEA |
| Omicron | XAK | Germany | (y) | Jun 22 | No evidence | No evidence | No evidence | No longer detected. |
| x: A67V, Δ69-70, T95I, G142D, Δ143-145, N211I, Δ212, ins215EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F z: A67V, Δ69-70, Δ143-145, N211I, Δ212, G339D, S371F, S373P, S375F, D405N, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, D796Y, Q954H, N969K n/a: Not applicable, no WHO label has been assigned to this variant at this time. All sub-lineages of the listed lineages are also included in the variant, e.g. B.1.429.1 is included in B.1.427/B.1.429 as it is a sub-lineage of B.1.429. a: No assessment of transmission is given for variants in the monitoring category, only detected/not detected. b: The earliest detections from several different countries are close in time and there is no clearly demonstrated travel link to a specific country that explains the detections. c: The property of concern for this variant was the fact that there are reports of difficulties associated with detecting it in upper respiratory tract samples. These difficulties were not caused by primer-template mismatch but rather by the virus not being present in sufficient quantities in the upper respiratory tract. d: Any amino acid substitution. |
||||||||
Table 5: De-escalated variants.
Perspectives in the drug design for SARS-CoV-2 variants
The current SARS-CoV-2 genome site mutations are all subject to natural selection and drug screening, which reflect the adaptation of the virus to the treatments. Some mutations make the virus increase the infectivity and transmission, however, in terms of treatment, the previous drugs are still effective against SARS-CoV-2 variants [21]. Other mutations render part of the antibody ineffective against SARS-CoV-2 [22], and the virus escapes from immunity, which brings new challenges to treatment. Therefore, the exploration of broadly antiviral drugs is prior to being concentrated and developed. More importantly, the immunomodulatory and holistic therapy in host, including anti-inflammatory strategies and Chinese traditional medicine treatment, should be concerned in the drug design for SARS-CoV-2 variants.
The binding of SARS-CoV-2 S protein to the ACE2 receptor is a key activity for the virus to invade the human body, and most of the mutation sites that are currently being studied are located in the S protein [23]. Researchers need to test one by one to determine the impact of specific mutation sites on the life of the SARS-CoV-2 and then test whether the previous treatments such as monoclonal antibodies are still effective for the emerging variants. However, a SARS-CoV-2 variant often carries a combination of different mutation sites and becomes a multiple mutant strain, and the speed of mutation is rapid [24]. It is of high concern that the effects from different combinations of mutation sites will have on viral infection activities. As a result, the highly conservative sites of the SARS-CoV-2 genome as drug targets could be paid more attention to maintaining antiviral activities against variants.
Discussion
SARS-CoV-2 has caused a global pandemic since late 2019. With the increase of experience and knowledge, defensive measures and clinical treatment plans are adopted immediately according to observation and management of COVID-19 pandemic. Meanwhile, researchers are exploring more possibilities of repurposing drugs against SARS-CoV-2 [25]. However, the development of specific drugs and vaccines requires fundamental studies on SARS-CoV-2, such as the interaction between the virus and the host, the epidemiology and molecular virology, the host immune responses to viral infection, to uncover the mechanism underlying the infection, transmission, and pathogenesis of the virus and explore effective drugs.
Traditional Chinese medicine has played a huge role in China’s anti-epidemic process [26]. Research on the mechanism of action of traditional Chinese medicine may provide new ideas for the development of anti-SARS-CoV-2 drugs. At the same time, it is necessary to pullulate holistic treatments to deal with the systemic pathological imbalance caused by the SARS-CoV-2.
Owing to the error-prone RNA replicase of SARS-CoV-2 the continuous emergence of mutant strains has undoubtedly brought huge difficulties and challenges to the control of the global COVID-19 epidemic [27]. The appearance of SARS-CoV-2 variants means that the virus adapts to manual interventions and natural selection, which may eliminate the effectiveness of previous drugs and vaccines. In this case, we must maintain epidemiological surveillance of COVID-19 timely and focus on the evolution of the virus, especially the emergence of drug-resistant strains.
The pathogenesis of COVID-19 can be mainly divided into two phases [28]. In the early stage, the COVID-19 patients may begin with a series of mild symptoms including fever, cough, fatigue, hemoptysis, headache, or diarrhea, and then will develop pneumonia [29]. Accordingly, antiviral drugs can be given to clear the virus from the body of patients infected with SARS-CoV-2. Later on, as the viral load increases and spreads in the human body, which triggers excessive production of cytokines, a cytokine storm occurs in the COVID-19 patients to deteriorate to moderate and severe symptoms with respiratory distress syndrome and organ failure. It should be necessary to adopt antiviral drugs with immunomodulatory treatment, and also encouraged to introduce holistic therapy and traditional Chinese medicine treatment, and even external mechanical equipment and maintain the normal operation of the patient’s body.
Conclusion
SARS-CoV-2 mutation is an urgent issue that needs to be explored and solved, which requires concerted efforts from the fields of structural biology, medicine, virology, pharmacy, public health, epidemiology, chemistry, and other disciplines, to jointly deal with the treatment and control of COVID-19 pandemic.
In patients at high risk, treatment with mAbs alone might not provide a therapeutic benefit in regions of the globe in which BQ.1.1 is spreading, suggesting that additional treatment options (e.g. paxlovid or molnupiravir) should be considered. Furthermore, novel, broadly active mAbs are urgently needed for prophylactic or therapeutic treatment, or both, in patients at high risk.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflict of Interest
The authors do not have a commercial or other association that might pose a conflict of interest.
Financial Support
None.
References
- Yin J, Li C, Ye C, Ruan Z, Liang Y, Li Y, et al. Advances in the development of therapeutic strategies against COVID-19 and perspectives in the drug design for emerging SARS-CoV-2 variants. Comput Struct Biotechnol J. 2022;20:824-837.
[Crossref] [Google Scholar] [PubMed]
- Network HR, Thompson MG, Yoon SK, Naleway AL, Meece J, Fabrizio TP, et al. Association of mRNA vaccination with clinical and virologic features of COVID-19 among US essential and frontline workers. JAMA. 2022;328(15):1523-1533.
[Crossref] [Google Scholar] [PubMed]
- Olson SM, Newhams MM, Halasa NB, Price AM, Boom JA, Sahni LC, et al. Effectiveness of BNT162b2 vaccine against critical COVID-19 in adolescents. N Engl J Med. 2022;386(8):713-723.
[Crossref] [Google Scholar] [PubMed]
- Milam AN, Doan DT, Childress DT, Durham SH. evaluation of monoclonal antibodies in preventing hospitalizations, emergency department visits, and mortality in high-risk COVID-19 patients. J Pharm Technol. 2022;38(3):169-173.
[Crossref] [Google Scholar] [PubMed]
- Levin MJ, Ustianowski A, De Wit S, Launay O, Avila M, Templeton A, et al. Intramuscular azd7442 (tixagevimab-cilgavimab) for prevention of COVID-19. N Engl J Med. 2022;386(23):2188-2200.
[Crossref] [Google Scholar] [PubMed]
- Arora P, Kempf A, Nehlmeier I, Schulz SR, Jack HM, Pohlmann S, et al. Omicron sublineage BQ. 1.1 resistance to monoclonal antibodies. The Lancet Infect Dis. 2023;23(1):22-23.
[Crossref] [Google Scholar] [PubMed]
- Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JC, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med. 2020;217(11):e20201181.
[Crossref] [Google Scholar] [PubMed]
- Venturelli S, Benatti SV, Casati M, Binda F, Zuglian G, Imeri G, et al. Surviving COVID-19 in Bergamo province: A post-acute outpatient re-evaluation. Epidemiol Infect. 2021;149.
[Crossref] [Google Scholar] [PubMed]
- Al-Aly Z, Xie Y, Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594(7862):259-264.
[Crossref] [Google Scholar] [PubMed]
- Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595(7866):283-288.
[Crossref] [Google Scholar] [PubMed]
- Lou JJ, Movassaghi M, Gordy D, Olson MG, Zhang T, Khurana MS, et al. Neuropathology of COVID-19 (neuro-COVID): clinicopathological update. Free neuropathol. 2021;2:2.
[Crossref] [Google Scholar] [PubMed]
- Steinman JB, Lum FM, Ho PP, Kaminski N, Steinman L. Reduced development of COVID-19 in children reveals molecular checkpoints gating pathogenesis illuminating potential therapeutics. Proc Natl Acad Sci. 2020;117(40):24620-24626.
[Crossref] [Google Scholar] [PubMed]
- Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, et al. Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature. 2008;451(7182):1076-1081.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Wang R, Gilby NB, Wei GW. Omicron variant (B. 1.1. 529): Infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62(2):412-422.
[Crossref] [Google Scholar] [PubMed]
- Dampalla CS, Zheng J, Perera KD, Wong LY, Meyerholz DK, Nguyen HN, et al. Post infection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc Natl Acad Sci USA. 2021;118(29):e2101555118.
[Crossref] [Google Scholar] [PubMed]
- Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, et al. Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. Cell Host Microbe. 2021;29(5):747-751.
[Crossref] [Google Scholar] [PubMed]
- Collier DA, de Marco A, Ferreira IA, Meng B, Datir R, Walls AC, et al. SARS-CoV-2 B. 1.1. 7 sensitivity to mRNA vaccine-elicited convalescent and monoclonal antibodies. MedRxiv. 2021.
[Crossref] [Google Scholar] [PubMed]
- Sun D, Sang Z, Kim YJ, Xiang Y, Cohen T, Belford AK, et al. Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nature Commun. 2021;12(1):4676.
[Crossref] [Google Scholar] [PubMed]
- McCallum M, de Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(9):2332-2347.
[Crossref] [Google Scholar] [PubMed]
- Hossain MU, Bhattacharjee A, Emon M, Hossain T, Chowdhury ZM, Ahammad I, et al. Novel mutations in NSP-1 and PLPro of SARS-CoV-2 NIB-1 genome mount for effective therapeutics. J Genet Eng Biotechnol. 2021;19(1):1-0.
[Crossref] [Google Scholar] [PubMed]
- Majumdar P, Niyogi S. SARS-CoV-2 mutations: The biological trackway towards viral fitness. Epidemiol Infect. 2021;149.
[Crossref] [Google Scholar] [PubMed]
- Han N, Hwang W, Tzelepis K, Schmerer P, Yankova E, MacMahon M, et al. Identification of SARS-CoV-2-induced pathways reveals drug repurposing strategies. Sci Adv. 2021;7(27):eabh3032.
[Crossref] [Google Scholar] [PubMed]
- Yang Y, Islam MS, Wang J, Li Y, Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): A review and perspective. Int J Biol Sci. 2020;16(10):1708.
[Crossref] [Google Scholar] [PubMed]
- Castro C, Arnold JJ, Cameron CE. Incorporation fidelity of the viral RNA-dependent RNA polymerase: A kinetic, thermodynamic and structural perspective. Virus research. 2005;107(2):141-149.
[Crossref] [Google Scholar] [PubMed]
- Tian W, Zhang N, Jin R, Feng Y, Wang S, Gao S, et al. Immune suppression in the early stage of COVID-19 disease. Nat Commun. 2020;11(1):5859.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63:364-374.
[Crossref] [Google Scholar] [PubMed]
- Xie J, Ding C, Li J, Wang Y, Guo H, Lu Z, et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J Med Virol. 2020;92(10):2004-2010.
[Crossref] [Google Scholar] [PubMed]
- Ucciferri C, Vecchiet J, Falasca K. Role of monoclonal antibody drugs in the treatment of OVID-19. World J Clin Cases. 2020 8(19):4280.
[Crossref] [Google Scholar] [PubMed]
- Pham T, Brochard LJ, Slutsky AS. Mechanical ventilation: State of the art. Mayo Clin Proc. 2017;92(9):1382-1400.
[Crossref] [Google Scholar] [PubMed]
Citation: Weimer LE, Cattari G, Binelli A, Belasio EF, Piras S, Sensi F (2023) Lastest Update on Therapeutics Strategies for the New Emerging COVID-19 Variants: A Review. Clin Microbiol. 12:353.
Copyright: © 2023 Weimer LE, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
