Indexed In
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Scimago
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2021) Volume 12, Issue 10
Isolation and Characterization of Siderophore and HCN Producing Pseudomonas aeruginosa from Indian Pompano Trachinotus mookalee Cuvier, 1832
Anu Mathew*, Imelda Joseph and Susmitha VReceived: 23-Sep-2021 Published: 12-Oct-2021, DOI: 10.35248/2155-9546.21.12.659
Abstract
Bacterial diversity study in marine fin-fish, Trachinotus mookalee Cuvier, 1832 lead to the isolation of six strains of Pseudomonas spp. The bacteria were isolated from the skin, gills and intestine of live fish collected from Karwar, Karnataka, India (N- 13°, 05.722‘; E- 079°, 48.658‘). Among the six strains three were non-fluorescent and three fluorescent Pseudomonas strains. Fluorescent strains were found to secrete pyoverdine, a yellow-green siderophore which has wide application in agriculture and allied sectors for use as biocontrol agent. Fluorescent strains were characterized phenotypically as well as by 16S rdna sequencing and were identified as Pseudomonas aeruginosa strain TRG1 (Genbank Accession No. KC109784).
Keywords
Fluorescent bacterium; Biocontrol; Pseudomonas aeruginosa; Indian Pompano
Introduction
Pseudomonas is part of a large heterogeneous and ubiquitous group of microorganisms generally referred to as pseudomonads. They are characterized as being highly metabolically versatile, bioactive and prolific colonizers on different surfaces. They are gram-negative, straight or slightly curved rods with polar flagella. Pseudomonads are chemo-organotrophs with a respiratory, non-fermentative type of metabolism and are usually catalase and oxidase positive. They are medically, environmentally and biotechnologically important bacteria. Pseudomonads are characterized by its enormous metabolic capacity, which is reflected by their capability to adapt to diverse and challenging environments, degrade recalcitrant compounds, and synthesize a variety of low-molecular weight compounds as well as biopolymer. It has been reported that pseudomonads constitute a large part of the microflora of the skin, gills and intestinal tracts of live fish [1,2] and are rarely reported as pathogens of fish [3]. Pseudomonas has been identified to be of importance in bioremediation as a result of their tremendous capacity for biodegradation. They also offer considerable promise in agronomic applications, since many strains are bioactive, fast-growing, prolific colonizers of plant surfaces and are able to suppress or out-compete pathogenic and other deleterious microorganisms [4]. The genus Pseudomonas is capable of using different substrates, such as glycerol, mannitol, fructose, glucose, n-paraffins and vegetable oils to produce rhamnolipid-type bio-surfactants [5,6]. Rhamnolipids produced by strains of Pseudomonas aeruginosa were shown to be highly effective against plant pathogens, including Pythium aphanidermatum, Plasmoparalactucae-radicis and Phytophthora capsci [7]. Fluorescent Pseudomonas spp. secretes pyoverdine, a fluorescent yellow-green siderophore under iron-limiting conditions. Such species have been used as biocontrol agents in several rhizosphere studies, where their inhibitory activity has been attributed to factors like production of antibiotics, hydrogen cyanide, and iron-chelating siderophores [8]. Pseudomonas fluorescens strains were applied in protecting roots of some plant species against parasitic fungi such as Fusarium or Pythium, as well as some phytophagous nematodes [9,10]. Pseudomonas aeruginosa has been reported as a biological control agent against plant pathogenic fungus Sclerotina sclerotiorum, a pathogen of more than 400 plant species around the world [11]. The renewed interest in biocontrol is due to its environmental friendliness, a long lasting effect and safety features. Some of the bacterial antagonists have also been found to show direct growth promoting effect on crop plants [12,13].
The present study was aimed to isolate and characterize bacterial strains of agricultural importance from economically important marine finfishes. Indian pompano Trachinotus mookalee Cuvier, 1832 is a demersal carangid fish with good flesh quality and mariculture potential.
Materials and Methods
Sample collection and processing
Live Indian pompano Trachinotus mookalee Cuvier, 1832were collected from shore seines at Karwar, Karnataka, India (N- 13°, 05.722‘; E- 079°, 48.658‘). For bacterial screening, the fish were anaesthetized and transported to the laboratory. For gut isolates, the fish were dissected under aseptic condition and the alimentary canals were removed. The alimentary canals from individual fish were cleaned, cut into pieces and by a longitudinal incision the contents were transferred to sterile petridish and flushed with chilled sterile saline (pH 7.4; 0.89 % (w/v) sodium chloride). Further, the parts of the alimentary canal were also homogenized in sterile saline (10:1; volume: weight). The homogenate thus obtained was used as inoculum for bacterial culture. For skin isolates, mucus extracts were isolated from the skin by swabbing aseptically. For gill isolates, the gills were macerated in 50 ml normal saline and centrifuged aseptically and the supernatant was used as inoculum for microbial culture.
Isolation and culture of bacterial strains
The samples were subjected to 10-fold serial dilution and aliquots (0.1 ml) were plated in duplicates onto nutrient agar supplemented with 2 % (w/v) sodium chloride. After 48 h of incubation at 35°C, well isolated colonies with distinct morphological characteristics were streaked onto nutrient agar plates to obtain pure culture and were stored at 4 °C for further studies. Nutrient broth with 2 % (w/v) NaCl and nutrient agar medium (HiMedia, Mumbai) with 2 % (w/v) NaCl was used to maintain the bacterial strains, to prepare the inocula and as the basal medium for phenotypic tests. King’s B medium (composition: protease peptone: 20 g L-1, K2HPO4: 1.5 g L-1, MgSO4.7H2O: 1.5 g L-1, agar: 20 g L-1, glycerol: 10 g L-1; at pH 7.2 ±1 and 35 °C) specific for fluorescent bacteria was used for culture and maintenance of fluorescent Pseudomonas spp.
Characterization
Phenotypic characterization: Holt JG, et al. [14] were followed for phenotypic characterization of bacterial strains and young cultures of 18- 24 h in nutrient broth and nutrient agar were used. Fluorescence was examined under UV light using 48 h old culture grown on King’s B medium. Growths at different concentrations of salt were determined by inoculating the bacteria in nutrient broth containing 0, 5, 9 and 12 % (w/v) sodium chloride (NaCl). The growth at pH 5, 9 and 10 was also determined by pH adjusted using 1N hydrochloric acid (HCl) or sodium hydroxide (NaOH). Biochemical tests carried out include catalase, cytochrome oxidase, penicillin sensitivity, H & L glucose O/F, sugar fermentation tests, cellulose hydrolysis, gelatin liquefaction, starch utilization and phosphate solubilization test.
Genotypic characterization: The pure cultures of fluorescent strains in tryptone soya broth (TSB, Oxoid) for 48 h were centrifuged for 10 min at 5031 × g. A total of 10–20 mg of each bacterial culture was placed in a 1.5 ml microcentrifuge tube and re-suspended with 200 μl of TE buffer. Bacterial DNA was extracted using Genomic DNA Purification Kit (Genie, Bangalore, India).
Amplification of partial 16S rDNA gene sequences and phylogenetic analysis
Identification of the strains was performed by sequence analysis of DNA coding for the 16S rDNA. Universal bacterial 16S rDNA primers were used to amplify a fragment of 16S rDNA of 760 bp in length. The PCR reaction mixtures contained 10 × PCR buffer with (NH4)2SO4, 2 mM dNTP Mix, 1.5 mM MgCl2, 20 pmol of each primer, 1 ng of DNA in 10 μL, and 2 Units of Taq DNA polymerase in a total volume of 50 μL. The PCR reactions were performed using initial denaturation during 3 min at 95°C followed by 30 cycles of denaturation for 1 min at 95°C, primer annealing for 1 min at 60°C, and primer extension for 1 min at 72°C. This procedure was followed by a final extension reaction at 72°C for 10 min. For negative controls for PCR reactions, instead of DNA, sterile distilled water was used. The PCR product was bi-directionally sequenced using the forward, reverse and internal primers. The multiple sequence alignment program Clustal W [15] was used to align the 16S rDNA sequence of the strains. Sequences of rDNA genes, for comparison, were obtained from the NCBI Gen Bank and RPD data base. Evolutionary distance matrices were calculated by using the algorithm of Kimura two-parameter model [16]. Phylogenetic tree was constructed by using the neighbour-joining method [17] with bootstrap re-sampling (data re-sampled 100 times) to assess the degree of support for the phylogenetic branching indicated by the optimal tree.
Assay for siderophore and hydrogen cyanide (HCN) production
The bacteria were streaked on the centre of Chrome Azurol S (CAS) agar media and incubated at 28 ± 2°C for 48h. When the bacteria consume iron, present in the blue-colored CAS media, orange halos around the colonies indicate the incidence of siderophores [18]. The production of HCN was detected by spreading 1 ml of 24 h old broth culture of Pseudomonas on the Kings B medium and incubation of the plates with the Whatman filter paper flooded with the solution containing 0.5% picric acid in 2% sodium carbonate located in the upper lid of petri plate [19]. To avoid the escape of the gas, the plates were sealed with the parafilm. After 24-48 h, yellow to orange change in the color of the filter paper was observed.
Results and Discussion
Increased concern over the impact of chemical fungicide on the environment has resulted in the increased interest in biocontrol strategies for the management of pathogens in agriculture and allied sectors. Out of the six strains identified as Pseudomonas spp., three were fluorescent strains, which secreted a yellow-green siderophore on King’s B medium (Figures 1 and 2). The biochemical characters of the strains are given in (Table 1). The strains were Gram-negative, motile short rods. The colonies on nutrient agar were light brown, translucent, oval or circular, flat or convex with smooth and shiny texture. They were positive to catalase, oxidase and citrate tests. It was non-sporulating, non-lactose fermenting and showed penicillin resistance. The tests were negative for indole, methyl red, Voges-Proskauer, starch utilization and cellulose hydrolysis and were able to reduce nitrate. The morphological, physiological and biochemical characteristics of the strains are almost similar, as reported in (Table 1). Fluorescent strains showed oxidative type of metabolism in H&L medium, whereas each of the non-fluorescent strain showed different reactions like oxidative, alkaline and no change. Based on the results, the three fluorescent strains were identified as Pseudomonas aeruginosa.
| Characteristics | Fluorescent Strains | Non Fluorescent Strains |
|---|---|---|
| Morphological | ||
| Light brown, translucent, oval or circular, flat or convex colonies on nutrient agar with smooth and shiny texture | ||
| Biochemical | ||
| Gram-negative, motile, short rods | ||
| Catalase | + | + |
| Oxidase | + | + |
| Citrate utilization | + | + |
| Fluorescence | + | - |
| Spore forming | - | - |
| Lactose fermenting | - | - |
| Indole | - | - |
| Methyl Red | - | - |
| Voges-Proskauer | - | - |
| Starch Utilization | - | - |
| Cellulose hydrolysis | - | - |
| Gelatin liquefaction | + | + |
| Penicillin sensitivity | Resistant | Resistant |
| Nitrate reduction | + | + |
| Oxidation fermentation | Oxidative | Oxidative/alkaline |
| Physiological | ||
| Growth in NaCl (%) | 0-5% | 0-9% |
| Growth in different pH | 5.0-10.0 | 5.0-10.0 |
Table 1: Phenotypic characteristics of fluorescent and non-fluorescent Pseudomonas spp. isolated from Trachinotus mookalee Cuvier, 1832.
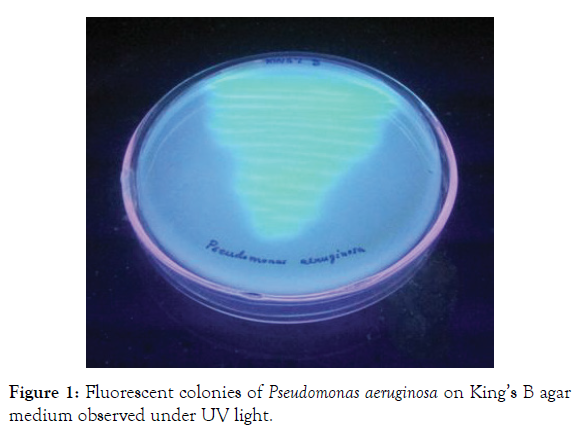
Figure 1: Fluorescent colonies of Pseudomonas aeruginosa on King’s B agar medium observed under UV light.
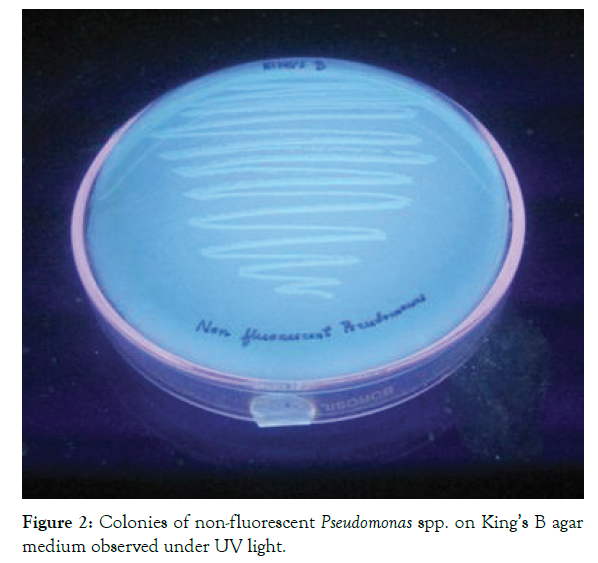
Figure 2: Colonies of non-fluorescent Pseudomonas spp. on King’s B agar medium observed under UV light.
Genotypic characterization
The 16S rDNA gene sequence of the three strains were aligned and compared with all sequences currently available for members of the genus Pseudomonas and related taxa (Figure 3) by using the NCBI Basic Local alignment search tool BLASTn program (http://www.ncbi.nlm.nih.gov/BLAST). Based on the nucleotide homology and phylogenetic analysis all three were found to be the same and was identified as Pseudomonas aeruginosa strain TRG1 (Genbank Accession No. KC109784). Also, the colour of CAS agar was changed from blue to yellowish orange confirming the production of siderophore. It has been reported that the siderophore producing microorganisms suppress many fungal pathogens through direct role of siderophore-mediated iron competition in the biocontrol ability [20]. Microbial production of HCN has been suggested as an important antifungal feature to control root fungi pathogen [21]. Cyanide acts as a general metabolic inhibitor to avoid predation or competition. The host plants are generally not harmfully affected by inoculation with HCN production bacteria and host-specific rhizobacteria can operate as biological control agents [22]. Unlike chemical pesticides, any biocontrol agent effectively colonizes in plant rhizosphere and shows long term effect in soil ecosystem. It has been reported that under certain condition, antibiotics improve the ecological fitness of these rhizobacteria in rhizoshpere, which can further influence long-term biological control efficacy [23]. P. aeruginosa are also able to produce compounds of industrial interest like enzymes (alkaline protease, lipase, chitinase, thermostable keratinase etc.), biosurfactants, biocatalyst for biofuel cell application, polymers (exopolysaccharides, poly-hydroxyalkanoates) etc [24]. Rhamnolipids, produced by Pseudomonas aeruginosa, represent an important group of biosurfactants having various industrial, environmental, and medical applications [25]. The role of Peudomonas aeruginosa in marine fish has not yet been established and further studies are necessary in this aspect. Since this strain has application in agriculture as a biocontrol agent for crops, its role in fish biochemistry cannot be ignored, this needs further research into it.
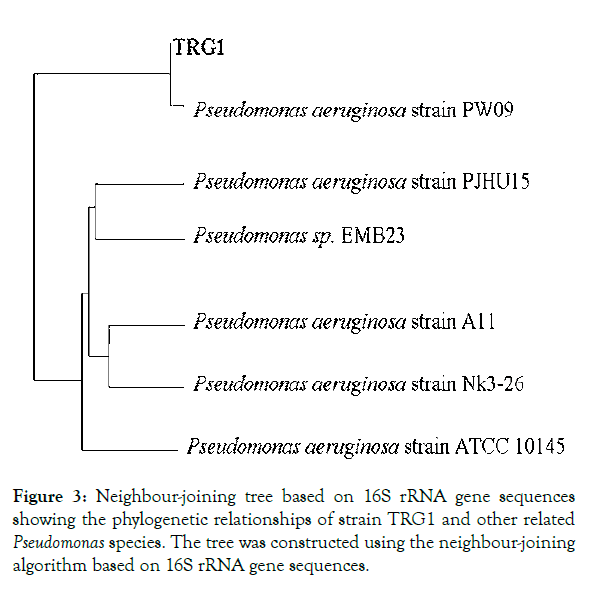
Figure 3: Neighbour-joining tree based on 16S rRNA gene sequences showing the phylogenetic relationships of strain TRG1 and other related Pseudomonas species. The tree was constructed using the neighbour-joining algorithm based on 16S rRNA gene sequences.
Acknowledgement
The authors acknowledge Indian Council of Agriculture (ICAR), New Delhi, India for funding of the work through the National Network Project Application of Microorganism in Agriculture and Allied Sectors (AMAAS). The authors are grateful to the Director, Central Marine Fisheries Research Institute (CMFRI), Cochin, Kerala, India for providing the facilities to carry out this work.
REFERENCES
- Cahill MM. Bacterial flora of fishes: A review. Microb. Ecol. 1990;19:21–41.
- Strøm E, Ringø E. Changes in bacterial flora in developing cod, Gadus morhua (L.), larvae after inoculation of Lactobacillus plantarum in the water. Physiological and biochemical aspects of fish larval development. 1993;226-228.
- Hatai K. Saprolegnia parasitica from rainbow trout inhibited by bacterium Pseudomonas fluorescens. Bull Eur Ass Fish Pathol. 1988;8:27-29.
- Nakai T, Hanada H, Muroga K. First records of Pseudomonas anguilliseptica infection in cultured ayu, Plecoglossus altivelis. Fish Pathol. 1985;20:481-484.
- Desai JD, Banat IM. Microbial production of surfactants and their commercial potential. Microbiol. Mol Biol Rev. 1997;61: 47−64.
- Koch AK, Käppeli O, Fiechter A, Reiser J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants.J Bacteriol. 1991;173:4212-4219.
- Gumaa NH. Inhibitory activity of Rhamnolipid produced by Pseudomonas aeruginosa NH22 against Rhizoctonia solani. Res J Biotechnol. 2012;6:32-44.
- O'sullivan DJ, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662-676.
- Chigusa K, Hasegawa T, Yamamoto N, Watanabe Y. Treatment of wastewater from oil manufacturing plant by yeasts. Water Sci Technol. 1996;34:51-58.
- Dong H, Gao S, Han SP, Cao SG. Purification and characterization of a Pseudomonas spp. lipase and its properties in non-aqueous media. Biotechnol Appl Biochem. 1999;30:251-256.
- Deshwal VK. Pseudomonas aeruginosa as biological control agent against plant pathogenic fungus Sclerotina sclerotiorum. J Anim Plant Sci. 2012;2:14-17.
- Glick BR, Karaturovic DM, Newell PC. A novel procedure for rapid isolation of plant growth promoting pseudomonas. Can J Microbiol. 1995;41:533-536.
- Deshwal VK, Pandey P, Kang SC, Maheshwari DK. Rhizobia as a biological control agent against soil borne plant pathogenic fungi.
- Holt JG, Krieg NR, Sneath PH. Bergey's manual of determinative bacterology.
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res Spec Publ.2003;31:3497-3500.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111-120.
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-425.
- Gopalakrishnan S, Humayun P, Kiran BK, Kannan IG, Vidya MS, Deepthi K, et al. Evaluation of bacteria isolated from rice rhizosphere for biological control of charcoal rot of sorghum caused by Macrophomina phaseolina (Tassi) Goid. World J Microbiol Biotechnol. 2011;27:1313-1321.
- Gajbhiye A, Rai AR, Meshram SU, Dongre AB. Isolation, evaluation and characterization of Bacillus subtilis from cotton rhizospheric soil with biocontrol activity against Fusarium oxysporum. World J Microbiol Biotechnol. 2010;26:1187-1194.
- Chaiharn M, Chunhaleuchanon S, Lumyong S. Screening siderophore producing bacteria as potential biological control agent for fungal rice pathogens in Thailand. World J Microbiol Biotechnol. 2009;25:1919-1928.
- Ramette A, Moënne-Loccoz Y, Défago G. Prevalence of fluorescent pseudomonas producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol Ecol. 2003;44:35-43.
- Saharan BS, Nehra V. Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res. 2011;21:30.
- Shanahan P, O'Sullivan DJ, Simpson P, Glennon JD, O'Gara F. Isolation of 2, 4-diacetylphloroglucinol from a fluorescent pseudomonas and investigation of physiological parameters influencing its production. Appl Environ Microbiol. 1992;58:353-358.
- Hussain S, Arshad M, Saleem M, Khalid A. Biodegradation of α-and β-endosulfan by soil bacteria. Biodegradation. 2007;18:731-740.
- Maier RM, Soberon-Chavez G. Pseudomonas aeruginosa rhamnolipids: biosynthesis and potential applications. Appl. Microbiol. Biotechnol. 2000;54:625-33.
Citation: Mathew A, Joseph I, Susmitha V (2021) Isolation and Characterization of Siderophore and HCN Producing Pseudomonas aeruginosa from Indian Pompano Trachinotus mookalee Cuvier, 1832. J Aquac Res Dev. 12:659.
Copyright: © 2021 Mathew A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.