Indexed In
- Open J Gate
- Genamics JournalSeek
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 14, Issue 5
PPM1D Truncation-associated Overexpression of the Stress-related Protein NQO1 Confers Sensitivity to the Bioactivatable Drug IB-DNQ in Diffuse Intrinsic Pontine Glioma
Maxime Janin*Received: 26-Sep-2024, Manuscript No. CPECR-24-27009; Editor assigned: 30-Sep-2024, Pre QC No. CPECR-24-27009 (PQ); Reviewed: 14-Oct-2024, QC No. CPECR-24-27009; Revised: 21-Oct-2024, Manuscript No. CPECR-24-27009 (R); Published: 28-Oct-2024, DOI: 10.35248/2161-1459.24.14.440
Abstract
Diffuse Intrinsic Pontine Glioma (DIPG) is a very aggressive brainstem tumor with a dismal prognosis and a lack of effective treatments. This study identifies the differential overexpression of the stress-related protein NAD(P) H Quinone Dehydrogenase 1 (NQO1) in some patient-derived DIPG cell lines and tumors. Given the critical role of NQO1 in cellular stress response, the objective was to clarify the mechanisms underlying its regulation in DIPG, while also exploring the therapeutic potential of the NQO1-bioactivatable drug Isobutyl-deoxynyboquinone (IB-DNQ). Interestingly, the study of the mutational profiles of the cell lines indicated that truncation of PPM1D correlated with NQO1 overexpression. From a functional perspective, cellular models were utilized to resolve that PPM1D phosphatase regulates NQO1 expression through the dephosphorylation of Mouse Double Minute 2 homolog (MDM2) at serine 395, resulting in the increased rate of translation of NQO1. From a therapeutic perspective, IB-DNQ treatment showed an NQO1-dependent growth inhibition sensitivity in vitro and extended survival in an in vivo DIPG model. These results reveal a new regulation of NQO1 at the protein level in PPM1D- mutated DIPGs indicating a potential therapeutic approach.
Keywords
DIPG; PPM1Dtruncation; NQO1; Isobutyl-deoxynyboquinone
Introduction
Patients with paediatric DIPG have very poor survival, and no effective treatment for the disease is available. DIPG is the main paediatric brainstem neoplasm, accounting for approximately 80% of brainstem tumours. It is mostly diagnosed in children between 5 and 10 years of age, and more than 90% of patients die within 2 years from diagnosis [1,2]. DIPG originates in the pons, a critical part of the brainstem, and diffusely infiltrates this very sensitive region controlling vital functions such as the regulation of respiration or involuntary functions, making complete surgical resection unfeasible. Recurrent mutations in the genes encoding the H3.3 (gene H3F3A) and H3.1 (genes HIST1H3B and HIST1H3C) histone variants, resulting in lysine-to-methionine substitution at position 27 (K27M), occur in more than 85% of DIPGs [3,4]. Other mutations have also been described as main drivers of DIPG development such as mutations present in PPM1D (protein phosphatase, Mg2+/Mn2+ dependent 1D), Activin A Receptor type 1 (ACVR1), Phosphoinositide-3-Kinase Regulatory subunit 1 (PIK3R1) and protein p53 [3,4]. The unique biology and clinical features of DIPG make therapeutic approaches for other paediatric brain cancer ineffective. Despite numerous clinical trials, the median overall survival remains around 12 months after diagnosis, with radiotherapy providing a 3 months survival benefit [5]. Therefore, it is urge to increase our understanding of the biology of DIPG and identify new treatment strategies to tackle this paediatric cancer.
It is known that tumors, including gliomas, use different pathways to mitigate oxidative stress-associated cell death. This study demonstrates that in patient-derived DIPG cell lines and biopsies, PPM1D-truncation mediates dephosphorylation of MDM2 which upregulates the multifactorial stress protein NQO1. Most importantly, it was noted that DIPGs overexpressing NQO1 were very sensitive to the antiproliferative effect of the NQO1- bioactivatable substrate IB-DNQ, a drug that crosses the blood-brain barrier, and is well-tolerated in preclinical models [6-8]. This drug is reduced by NQO1 in a futile cycle that generates high amount of Reactive Oxygen Species (ROS), leading to cell death. Consequently, the level of NQO1 expression is directly proportional to the anti- tumour effect of this compound. Therefore, it is proposed to assess the effect of IB-DNQ in preliminary clinical assays as a candidate therapeutic approach against PPM1D-truncated DIPG. This strategy could prove beneficial for DIPG patients.
Materials and Methods
Cell lines
Human DIPG patient’ derived cell lines HSJD-DIPG-007; HSJD- DIPG-008; HSJD-DIPG-011; HSJD-DIPG-012; HSJD-DIPG-013 and HSJD-DIPG-014 were obtained from the Sant Joan de Deu paediatric hospital in Barcelona (Catalonia, Spain); SU-DIPG-VI was obtained from Dr. Michelle Monje’s laboratory in Stanford (Stanford Medicine; USA). Cell lines were all cultivated in a 1:1 mixed Neurobasal-A medium with DMEM/F12 medium supplemented with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer, sodium pyruvate MEM, Non-essential amino- acids, GlutaMax-I, antibiotic-antimycotic, B-27 supplement without vitamin A (all from ThermoFisher), human PDGF-AA, human PDGF-BB, recombinant human FGF, recombinant human EGF (all from PeproTech) and heparin solution 0.2% (Sigma). These cell lines were cultivated regarding to suppliers recommendations, grown and maintained in appropriate conditions (37°C, 5% CO2). All cell lines were tested for the absence of mycoplasma.
Expression analyses
For real time Quantitative Reverse Transcription Polymerase Chain Reaction (QRT-PCR) experiments, total RNA extraction was performed, or one sample of brain white matter, using the SimplyRNA kit (Promega) on a Maxwell RSC device (Promega). Three were converted to cDNA using the ThermoScript RT-PCR system (ThermoFisher) with oligo(dT) primers. For real time QRT- PCR experiments, total RNA was extracted on three independent cellular pellets from each cell line using the SimplyRNA kit (Promega) on a Maxwell RSC device (Promega). The extracted RNA was stored at -80°C. Three µg of DNase treated RNA were retrotranscribed using the ThermoScript RT-PCR system (ThermoFisher) with oligo(dT) primers. Quantitative PCR was performed using SYBR® Green PCR Master Mix (ThermoFisher) on a QuantStudio5 (AppliedBiosystems) with QuantStudioTM Design&Analysis Software v1.3.1, and GAPDH was used as housekeeping gene to enable normalization. Primers for RT-PCR are listed in Supplementary Table 1. For immunoblotting assays, cell pellets and brain white matter samples were extracted using Laemli buffer, sonication and 5 min at 95°C. 80 µg of total cellular proteins were loaded per lane of Sodium Dodecyl Sulfate-polyacrylamide gels. Proteins were transferred to nitrocellulose membranes, then membranes were blocked in 5% non-fat dry milk in Tris-Buffered Saline supplemented with Tween20 (TBS-T, Biorad) and incubated with primary antibodies diluted in milk for 60 min at reverse transcription or overnight at 4°C. Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h before developing with Enhanced Chemiluminescence (ECL, Biorad). For immunohistochemical analysis, paraffin- embedded sections of DIPG tumors were immunostained with the NQO1 antibody. Antibodies used in this study are described in Supplementary Table 1.
Drug assays
Half-maximal Inhibitory Concentration (IC50) studies were performed using the 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H- tetrazolium bromide (MTT) assay in drug-treated DIPG cell lines. Briefly, 48 h after exposure to nine increasing concentrations of the drug, 10 µL of diluted MTT was added to culture medium and incubated in culture incubator for 4 h. Then, 100 µL of lysis buffer was added to each well and the plates were incubated overnight in the incubator. The days after, the 540 nm-optical densities were determined using a microplate reader (Perkin Elmer Viktor 3). Drugs used in this study are listed in Supplementary Table 1.
DNA methylation analyses
DNA was extracted from cell lines and brain white matter pellets by the addition of 4.5 mL of lysis buffer (10 mM Tris, 5 mM EDTA, 100 mM NaCl, pH 8), and re-suspended with 500 µL of SDS 10% and 50 µg of proteinase K at 10 mg/mL. Samples were incubated overnight at 37°C. After addition of 2.5 ml of NaCl 5 M and vortex, samples were centrifuged at 4000 rpm during 15 min. Upper phases were collected and 5.6 mL of isopropanol were added to precipitate DNA. The DNA was recovered and washed by ethanol 70%. After re-suspension in water, all DNA samples were quantified by NanoDrop. The DNA methylation array used was the Infinium MethylationEPIC BeadChip (Illumina). Raw fluorescence intensity values were normalized with Illumina Genome Studio software (V2011.1) using ‘control normalization’ with background correction. Normalized intensities were then used to calculate DNA methylation levels (beta values).
Gene transfection
The cDNA sequences of NQO1, PPM1D or MDM2 were cloned either into the pLVX-IRES-ZsGreen1 or pLVX-IRES-TOMATO expression plasmid (Clontech Laboratories) between defined restriction sites. Lentiviruses containing this construct were produced by cotransfecting HEK-293T cells with the recombinant plasmid, with packaging vectors and jetPRIME® Transfection reagent (Polyplus Transfection) following the supplier instructions. Briefly, 10 µg of each encoding plasmid was mixed with 7.5 µg of ps-PAX2 and 2.5 µg of PMD2.G plasmid (Addgene). Upon 10 min of RT incubation, the transfection mix was added dropwise on a 10 cm culture plate containing HEK293-TLV lentiviral packaging cells at 80% confluence. The transfection cocktail was removed after 6 h and replaced by fresh medium. After 72 h, viral-containing supernatant was collected, 0.45 µM-filtred and stored at 4°C before infection. The recombinant product was randomly inserted by lentiviral transduction in the genome of the DIPG cell lines. After 5 passages, the green or red fluorescent cells were sorted by Fluorescence Activated Cell Sorting (FACS) and cultured in the above-described DIPG-specific medium. For primers sequences please refer to Supplementary Table 1.
Gene depletion
Four different sequence gene-specific short hairpin RNA molecules (shRNAs) for NQO1 mRNA were designed and transduced into DIPG cell lines (for sequences please refer to Supplementary Table 1. shRNA against the MSS2 yeast mRNA (not present in mammals) was used as a scramble control. All shRNA molecules were ligated into pLVX-shRNA2-ZsGreen plasmid (Clontech). Lentiviral particles were produced following the same steps than in the previous section. Infected cells were FACS-sorted after 5 passages and cultured in DIPG-specific medium.
Brain tumor xenografts
For tumor growth into the brainstem pons area, 1 × 106 of HSJD- DIPG-007 cells were inoculated into the pons of 10-9 week-old athymic Nude-Foxn1nu mice (Charles River Laboratories) for each treatment condition. All mouse experiments were approved by and performed according to the guidelines of the Xenopat company in agreement with the European Union and the Spanish national directives. The experiments were also approved by the local ethics committee. Globally, the “Principles of laboratory animal care” (NIH publication No. 86-23, revised 1985) were followed.
After 20 days, half of the mice were randomly chosen to be treated with IB-DNQ intravenously at 12 mg/kg every 2 days for 10 days (5 doses) while the other half were treated with vehicle hydroxypropyl beta-cyclodextrin (HPbCD).
Patients’ samples
Tumor samples were obtained from autopsies, freshly frozen, and collected at Sant Joan de Deu paediatric hospital, Stanford Monje Lab, and Washington D.C. paediatric hospital. A specific Material Transfer Agreement (MTA) was established between the different institutions, and the institutions declared written consent established with the family of the donor.
Results
Overexpression of the NQO1 protein in patient-derived DIPG cell lines
Oxidative stress-induced death of cancer cells is a barrier to tumor growth and transformed cells have developed various mechanisms to overcome this challenge [9]. One such mechanism involves the induction of proteins that regulate this oxidative stresses. In this regard, NQO1 is a multifunctional antioxidant that provides significant protection against cell death [10]. Overexpression of the stress-related protein NQO1 has been described in several cancer types, including pancreatic, lung, breast, thyroid, adrenal, ovarian, colon and corneal tumors [11-14]. Its high expression is usually associated with poor prognosis [15-17]. The molecular pathways that lead to NQO1 overexpression in human malignancies are not completely understood and so far, no study of NQO1 protein expression has been performed in paediatric brainstem tumors. Using as starting biological material six patient-derived DIPG cell lines (all of them H3K27M-mutated), analysis was conducted using Western-blot high expression of NQO1 in three cases (Figure 1a). The remaining three DIPG cell lines (Figure 1a) and control human astrocytes and brain white matter samples (Supplementary Figure 1a) showed low levels of the NQO1 protein. The different levels of NQO1 were not linked to distinct DNA methylation patterns in the NQO1 promoter CpG island (Supplementary Figure 1b), or in the NQO1 regulator NSUN5 (Supplementary Figure 1c) [6]. Importantly, this overexpression was not limited to in vitro conditions since primary autopsy DIPG samples confirmed detectable NQO1 overexpression by immunohistochemistry. One block, N1327, corresponded to the autopsy that led to the derivation of the HSJD-DIPG-008 cell line, and this tumor had one of the highest expressions (Figure 1b). Interestingly, the NQO1 RNA expression levels among the six DIPG cell lines were not statistically different (Figure 1c), suggesting that the translocation of the transcription factor NRF2 into the nucleus, which leads to an increase of RNA transcription of different stress-related genes, including NQO1, was not involved either [18]. Thus, protein translational or posttranslational regulation could be involved to generate the different NQO1 protein levels observed. To know if NQO1 protein upregulation was due to different protein stability, cycloheximide chase analysis was employed to assess protein degradation in the studied DIPG cell lines. Western blot analysis of NQO1 level did not show loss of protein stability upon cycloheximide treatment (Figure 1d), suggesting that distinct protein levels could be related to different rates of translation. Indeed, NQO1 protein levels showed similar variation in all cell lines following cycloheximide treatment, with a time-dependent increase. This suggests that distinct translation rates, rather than differences in protein degradation, are responsible for the observed differences in NQO1 levels (Figure 1).
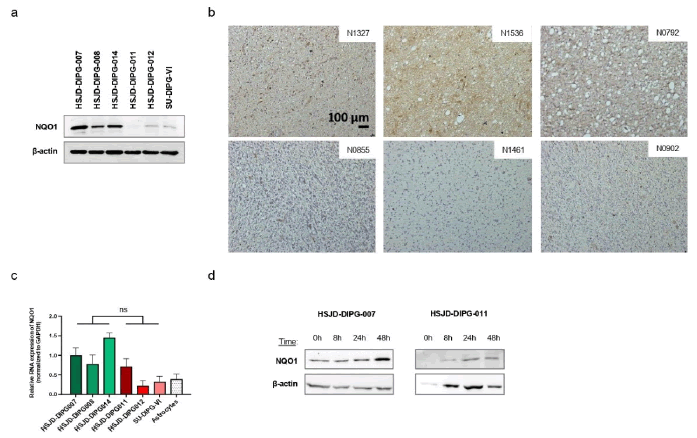
Figure 1: Expression of NQO1 in DIPG cell lines. Note: a) Western blot of NQO1 in the 6 DIPG cell lines; b) Representative images of immunodetection of NQO1 in 6 formalin-fixed paraffin embedded sections with same optical zoom and light exposure. Sections with highest expression of NQO1 are on the upper line and sections with lowest expression are on the line below. Section corresponding to HSJD-DIPG-008 cell line is N1327 up on the left; c) NQO1 expression levels in DIPG cell lines and astrocytes control determined by real-time PCR (data shown represent mean ± SD of biological triplicates); d) Western blot of NQO1 and beta-actin after cycloheximide treatment at 500 µM during different times in HSJD-DIPG-007, -008, -011 and -012; For real time PCR significance was calculated using an unpaired t-test; ns: not significant; All drug-response curves, real time PCR and relative expression bar plots were generated using GraphPad Prism software in this study.
PPM1D is mutated in DIPG cell lines with high expression of NQO1
The DIPG cell lines exhibited only slight differences in growth and physiological characteristics. All were morphologically similar and grew in the same culture medium, except for HSJD-DIPG-007, which demonstrated faster growth than the others. Nevertheless, to investigate which molecular factors might enhance translation rates, leading to NQO1 protein overexpression, it was evaluated the genetic context of the six studied DIPG cell lines beyond its shared H3K27M-mutated status. Remarkably, the 3 DIPG cell lines with high NQO1 protein expression levels were mutated for the PPM1D gene (also known as WIP1) and the other three, with low NQO1 protein expression levels, were wild type (Table 1) [19]. Moreover, the expression of NQO1 in PPM1D mutated freshly frozen autopsy tumours was higher than wild-type DIPG cases or normal cortex (Figure 2a), further supporting the occurrence of NQO1 overexpression in the primary tumour. The normal cortex sample was matched from the same patient with the SU-DIPG-35 PPM1D-truncated tumor, confirming that NQO1 upregulation was specific to the cancer cells.
| Cell line | PPM1D | TP53 | H3F3A |
|---|---|---|---|
| HSJD-DIPG-007 | P428X | WT | K28M |
| HSJD-DIPG-008 | R429X | WT | K28M |
| HSJD-DIPG-014 | T483TX | WT | K28M |
| HSJD-DIPG-011 | WT | WT | K28M |
| HSJD-DIPG-012 | WT | R273C | K28M |
| SU-DIPG-VI | WT | E198X | K28M |
Table 1: Mutational status of the DIPG cell lines. Mutations of PPM1D, TP53 and H3F3A genes are shown for the 6 DIPG cell lines used in this study.
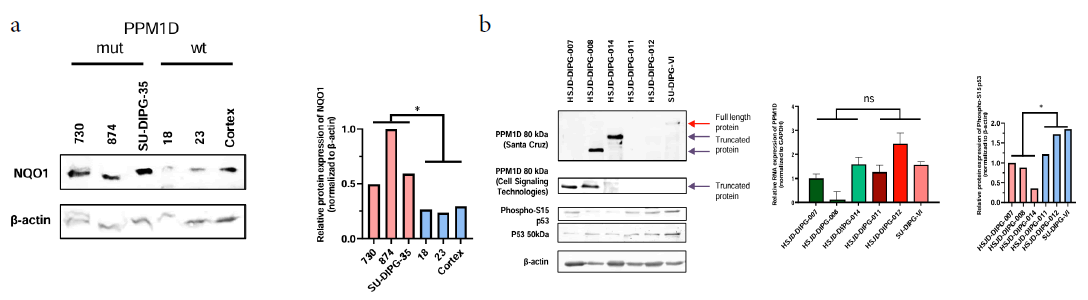
Figure 2: Patient-derived cell lines HSJD-DIPG-007; HSJD-DIPG-008 and HSJD-DIPG-014 are mutated for PPM1D. Note: a) Western blots of NQO1 in tumors from patients with or without PPM1D mutation; b) Left: Western blot of PPM1D showing accumulation in the 3 DIPG cell lines that overexpress NQO1 and western blot of p53 (S15) showing a slight variation between the two groups of cells; Middle: Real time PCR of PPM1D in the 6 DIPG cell lines studied; Right: Intensities of phospho-S15 p53 relative to beta-actin; Significance was calculated using an unpaired t-test; ns: not significant; *P<0.05.
PPM1D is mutated in approximately 20% of DIPG tumours, with point mutations in the sixth exon that result in protein truncation, leading to the production of C-terminal domain-lacking proteins [20]. The loss of this domain causes a protein accumulation that is still functionally active inside the cells, and impairs DNA damage response [21-23]. As expected, Western blot was performed using different antibodies depending on epitope recognition, the accumulation of PPM1D in the three mutant DIPG cell lines (Figure 2b). PPM1D RNA levels did not show differences among the cell lines (Figure 2b). Interestingly PPM1D dephosphorylates and destabilizes p53, while NQO1 stabilizes it [24-26]. Thus, PPM1D and NQO1 share a common target but with opposite activities and, thus, it was hypothesized that the observed NQO1 overexpression might represent a compensatory response by the cells against PPM1D accumulation and over activity. In this regard, it was found that a slight decrease of p53 protein levels in those DIPG cells overexpressing NQO1, but no clear variation of p53-S15 phosphorylation comparing with p53 protein level (Figure 2b), suggesting compensatory regulation both at p53 and p53 phosphorylation levels (Figure 2).
Mutant PPM1D regulates NQO1 expression in DIPG cell lines
To assess if PPM1D truncation had a direct effect on NQO1 expression, various genetically engineered cellular models of DIPG were developed (Supplementary Figure 2). It was noted that the stable transfection of the SU-DIPG-VI cell line with the PPM1D truncated form induced an increase in NQO1 protein levels (Figure 3). NQO1 upregulation was not observed upon transfection of the full-length form (Figure 3a), a mutant inert form of the full-length (Supplementary Figure 2a) or a mutant inert form of the truncated PPM1D protein (Supplementary Figure 2a). Conversely, investigated the loss of the truncated PPMD1 protein on NQO1 expression by generating a PPM1D knock-out cell model (Supplementary Figure 2b) by CRISPR-Cas9 in the PPM1D truncated mutant HSJD- DIPG-007 DIPG cells (Figure 3b). It was found that, upon efficient abrogation of PPMD1 expression, there was a reduction of NQO1 protein levels (Figure 3b). Interestingly, PPM1D inhibition by small molecules has been suggested as a possible treatment in DIPG to reactivate DNA damage response and decrease tumor growth [27]. Therefore, the effect of the PPM1D inhibitor GSK2830371 on NQO1 expression was evaluated in the PPM1D truncated mutant HSJD-DIPG-007 and HSJD-DIPG-014 cell lines, which resulted in decreased NQO1 expression (Figure 3c). Additionally, it was confirmed that the truncated form of PPM1D regulates NQO1 protein expression using a recently developed genetically modified mouse model [28]. It was determined that endogenous truncation of PPM1D exon 6 in brainstem tumors was associated with an increase in NQO1 protein levels (Figure 3d), but not in the corresponding RNA transcript (Figure 3d). In addition, the analysis of mouse Neural Stem Cell (mNSC) transfected with control Green Fluorescent Protein (GFP) or PPM1D-truncated plasmids demonstrated NQO1 overexpression at the protein level in PPM1D transfected cells, but not for the RNA transcript (Figure 3e).
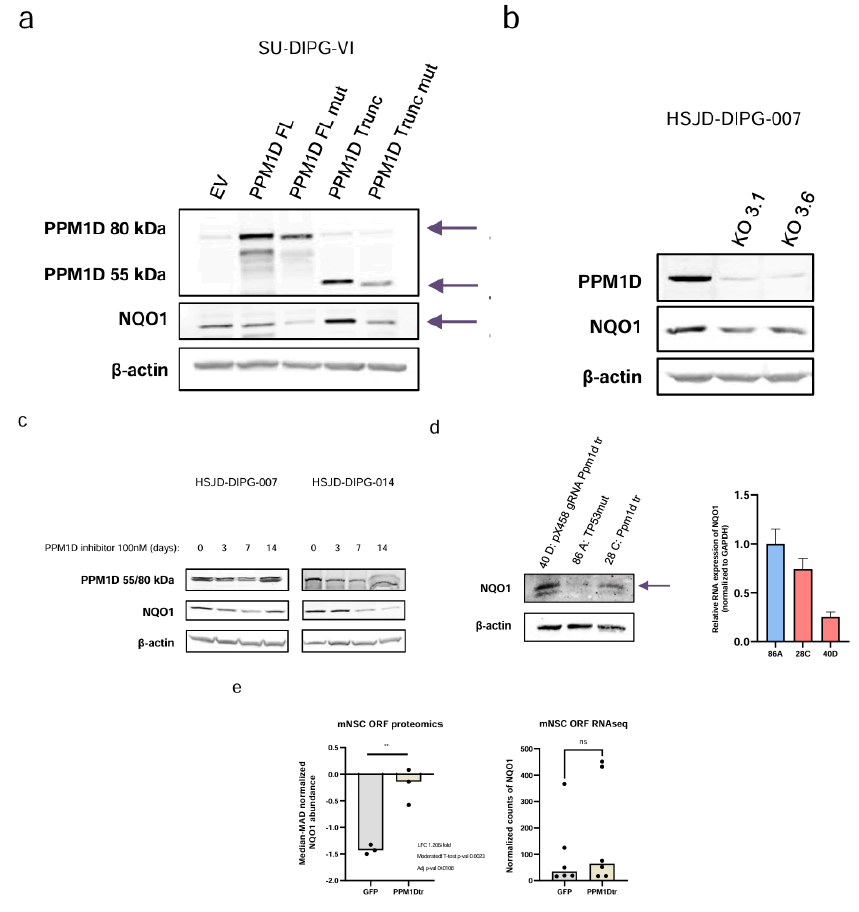
Figure 3: NQO1 expression is modulated by PPM1D. Note: a) Western blot analyses of PPM1D overexpression in SU-DIPG-VI cell line 6 weeks after stable transfection; b) Western blot of PPM1D and NQO1 in HSJD-DIPG-007 KO cell lines; c) Western blot of PPM1D and NQO1 in HSJD-DIPG-007 and HSJD-DIPG-014 cell lines treated at 100 nm during 0, 3, 7 and 14 days; d) NQO1 expression in mouse cell lines derived from IUE tumors analysed by western-blot (left) and real time PCR (right); e) Proteomic and RNA-seq expression of NQO1 in mouse Neural Stem Cells (mNSC) transfected with GFP as control or with PPM1D truncated; EV: Empty Vector; FL: Full Length; mut: Mutated; Trunc: Truncated; Arrow indicates the expected bands for PPM1D (full length or truncated) and NQO1; For (e) significance was calculated using a moderated t-test; ns: Not significant; **P<0.005.
Dephosphorylation of MDM2 at serine 395 by truncated PPM1D leads to NQO1 protein upregulation
These data suggest that the truncated form of PPM1D is a key regulator of NQO1 protein levels in DIPG. However, it remains unclear whether this regulation is a direct effect or mediated by downstream targets via PPM1D’s phosphatase activity. One of the most well-known targets of PPM1D is the oncoprotein MDM2 a negative regulator of p53 protein levels. In this regard, the mutations in the phosphatase PPM1D leads to dephosphorylation of MDM2 at the S395 residue; a change associated with MDM2 stabilization and enhanced interaction-induced degradation of p53 [29]. It was hypothesized that the PPM1D dependent-phosphorylation status of MDM2 could have additional roles beyond p53, for example acting through NQO1. To investigate this, a serine 395 to alanine MDM2 mutant plasmid was constructed for stable transfection in DIPG PPM1D wild-type cells. It was noted that the transfection of this S395A mutant that mimics the PPM1D phosphatase activity, in HSJD-DIPG-011 cells increased NQO1 protein expression level (Figure 4) but did not induce changes of NQO1 mRNA expression (Figure 4). It is possible that PPM1D-mediated dephosphorylation of MDM2 leads to NQO1 upregulation, a known p53 stabilizer [25]. This may help avoid excessive p53 degradation and cell death in DIPGs. In this regard, MDM2 regulates protein translation and this mechanism could be invoked herein, since changes in NQO1 protein degradation were not observed (Figure 1d) [30,31].
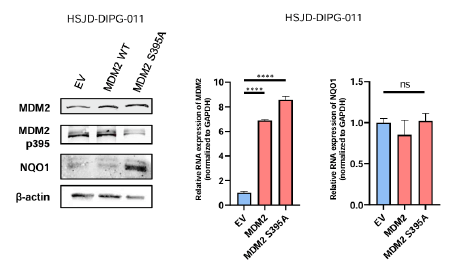
Figure 4: MDM2 serine dephosphorylation leads to NQO1 protein increased expression. Stable transfection in HSJD-DIPG-011 of MDM2 S395A validated by Western blot leads to NQO1 increased expression analysed by western blot (left), but do not modulate the transcript expression of NQO1 analysed by real time PCR (right). Note: For real time PCR, significance was calculated using an unpaired t-test; ns: Not significant; ****P<0.0001.
NQO1 overexpression in DIPGs with PPM1D truncation- mutations are sensitive to the bioactivatable drug IB-DNQ
The reduction of quinone substrates by NQO1 to their respective hydroquinones in a futile redox cycle results in a high production of Reactive Oxygen Species (ROS), causing extensive DNA lesions, PARP1 hyperactivation, and severe NAD+/ATP depletion that stimulate Ca2+–dependent programmed necrosis [32]. This mechanism proved useful in inducing an outsized oxidative stress level that leads cancer cells to their death [33]. In this regard, various drugs have been found to be NQO1-bioactivatable, such as Beta- lapachone (ARQ 501) or quinone derivates as deoxynyboquinone and isobutyl-deoxynyboquinone (DNQ and IB-DNQ, respectively), which have demonstrated their anticancer activity in cancer cell line models [33-35]. IB-DNQ has shown excellent pharmacokinetics and safe tolerability in domestic feline models that further support the preclinical development of this drug [7,8]. It was hypothesized that if those DIPGs harbouring PPM1D truncation-associated NQO1 overexpression showed a particular sensitivity to IB-DNQ.
Using the MTT assay to determine the IC50 cell growth inhibition values in my panel of DIPG cell lines, it was found that the 3 DIPG cell lines with PPM1D truncating mutation and associated high NQO1 expression (HSJD-DIPG-007; -008 and -014) were significantly more sensitive to the drug than the 3 DIPG cell lines with wild-type PPM1D and low levels of NQO1 (HSJD-DIPG-011; HSJD-DIPG-012 and SU-DIPG-VI) (z test, P<0.0001) (Figure 5a). The use of different NQO1 constructs in these cells further confirmed this link. The development of stable depletion models of NQO1 by transfection of short hairpin RNA (shRNA) in NQO1 high-expressor HSJD-DIPG-007 cells showed an acquired resistance to the growth inhibition effect mediated by IB-DNQ (Figure 5b). Conversely, the induction of NQO1 overexpression by stable transfection in cells with originally low-levels of NQO1 (HSJD-DIPG-011) rendered these cells to a more sensitive growth inhibitory effect upon IB-DNQ use (Figure 5c).
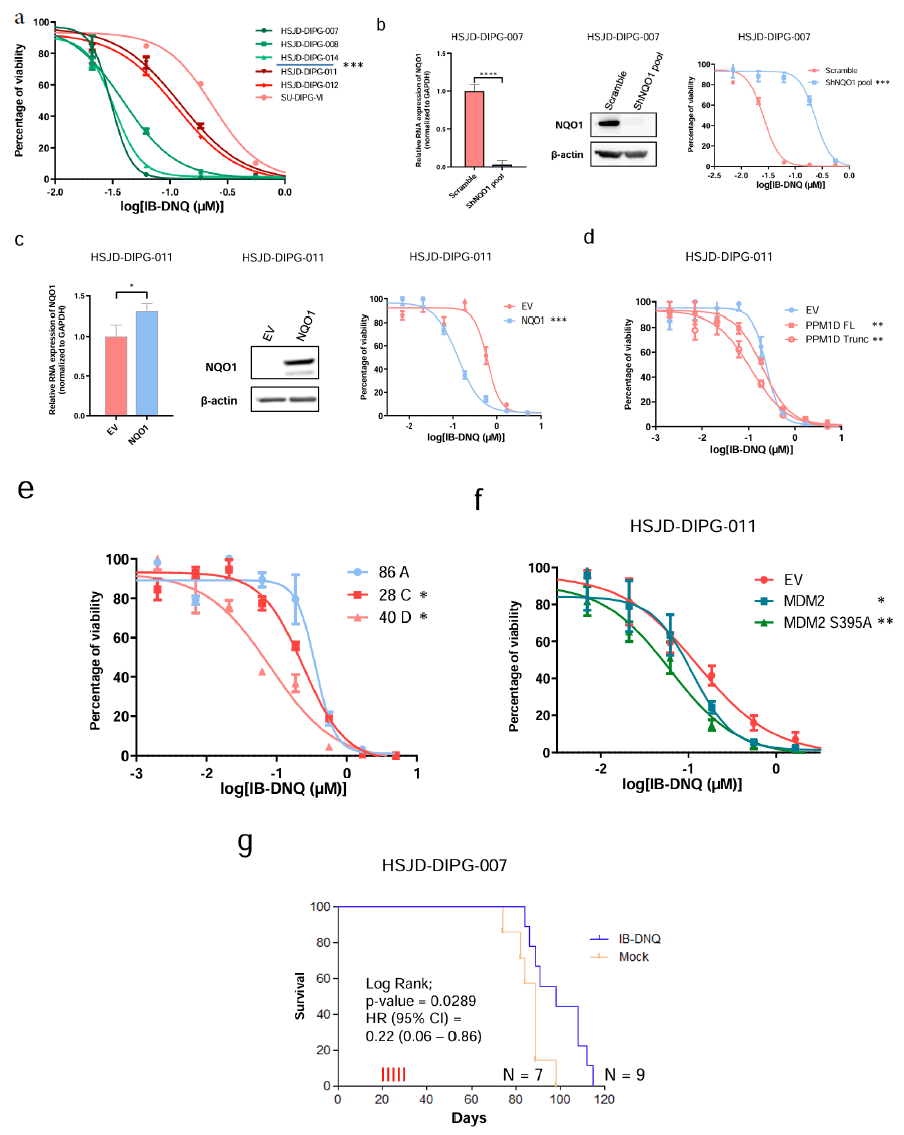
Figure 5: NQO1 modulation leads to IB-DNQ sensitivity in DIPG. Note: a) IC50 determination using the MTT assay in the DIPG cell lines; b) DIPG cellular models of NQO1 depletion by short hairpin (sh) RNA validated by real time PCR (left panel) and western blots (middle) for the cell line HSJD-DIPG-007; c) NQO1 overexpression model by stable transfection in HSJD-DIPG-011 cell line validated by real time PCR (left panel) and western- blots (middle) that leads to IB-DNQ sensitivity (right panel); d) PPM1D overexpression by stable transfection in HSJD-DIPG-011 cell line leads to IB- DNQ sensitivity; e) Cell lines derived from IUE mouse models of PPM1D are more sensitive to IB-DNQ; f) MDM2 and mainly MDM2 S395A stable transfection in HSJD-DIPG-011 lead to IB-DNQ sensitivity; g) Kaplan-Meier analysis of survival according treatment conditions (IB-DNQ treated vs. mock group) in a mouse model with implanted tumors derived from the NQO1 overexpressing DIPG cell line DIPG 7; *P<0.05; **P<0.005, ***P<0.0005; ****P<0.0001.
The involvement of the PPMD1 truncation mutation leading to NQO1 upregulation, and thus being itself a biomarker of response to the IB-DNQ compound, was also demonstrated for the herein described genetically engineered human and mice models. In this regard, it was noted that the stable transfection of the PPM1D truncated form in non-mutated DIPG cell lines not only induced an increase in NQO1 protein levels (Figure 3a), but also increased the sensitivity to IB-DNQ (Figure 5d). The use of the mouse model with a truncated form of PPM1D [28], demonstrated induced NQO1 overexpression, as shown previously (Figure 3d), further confirmed that the developed brainstem tumors were highly sensitive to IB- DNQ in comparison to non-truncated PPM1D specimens (Figure 5e). These observations were extended to the target of the PPM1D truncation, the oncoprotein MDM2. It was found that transfection of the S395A MDM2 mutant (that mirrors he PPM1D phosphatase activity) in DIPG PPM1D wild-type cells not only induced NQO1 upregulation (Figure 4), but also enhanced the sensitivity to the IB- DNQ drug (Figure 5f). As DIPG originates into the pons, a sensitive area into the brain that render difficult therapeutics’ delivery, the IB-DNQ treatment was evaluated in a mouse model with orthotopic xenograft [36]. In a first proof of principle experiment, the HSJD- DIPG-007 cell line was injected into the pons area of 16 mice. The results indicated that IB-DNQ treatment significantly increased the survival of mice compared to the control group (Figure 5g).
Overall, these data suggest that DIPGs carrying mutant truncated forms of PPM1D with associated overexpression of the stress related protein NQO1 are very sensitive to the anti-growth effect induced by the bioactivatable drug IB-DNQ, a molecular weakness that it is worth to be explored in further studies aimed to improve the survival of patients with currently dismal prognosis (Figure 5).
In Figure 5a, DIPG cells harbouring NQO1 overexpression (HSJD- DIPG-007; -008 and -014) show higher sensitivity to IB-DNQ in comparison to NQO1 normal expression cells (HSJD-DIPG-011; -012 and SU-DIPG-VI). In Figure 5b, NQO1 depletion leads to IB-DNQ resistance shown by IC50 determination (right panel). In Figure 5g, mice were treated at days 20, 22, 24, 26 and 28 (red vertical lines). Significance of the log-rank test is shown. Results of the univariate Cox regression analysis are represented by the Hazards Ratio (HR) and 95% Confidence Interval (CI). Significance was calculated using a paired t test for IC50s and an unpaired t-test for real time PCR.
Discussion
This study revealed NQO1 protein overexpression in DIPG patient- derived cell lines and primary tissues, and the strongest marker correlating with NQO1 overexpression was the presence of PPM1D truncation. Through various cellular models, it was demonstrated that PPM1D-targeted dephosphorylation of MDM2 at the S395 residue led to the accumulation of NQO1. However, additional studies are needed to precisely understand the interactions between p53, PPM1D, MDM2, and NQO1, and how these proteins influence one another in this network. Given that MDM2 has been shown to promote the translation of certain mRNAs, it is possible that this also occurs with NQO1 mRNA in this specific context.
The discovery that PPM1D mutations mediate NQO1 overexpression prompted the evaluation of the NQO1-bioactivatable compound IB-DNQ across different DIPG models. The drug exhibited potent activity against DIPG cell lines, and this activity was dependent on NQO1 overexpression driven by PPM1D truncation mutations. This suggests that DIPG patients possess PPM1D mutations could potentially benefit from IB-DNQ or similar compounds. However, extensive validation is needed to establish IB-DNQ as a viable treatment. Nevertheless, given the extremely poor prognosis of DIPG, these data provide an encouraging basis for more comprehensive preclinical studies that could prepare for future clinical applications. The use of NQO1-bioactivatable compounds may align well with existing treatment strategies for DIPG. For instance, several therapeutic agents, including radiotherapy (the current standard of care in DIPG) and chemotherapeutic agents like gemcitabine, doxorubicin, cisplatin, and vorinostat, have been reported to modulate NQO1 expression [37-42]. Thus, targeting NQO1 may not disrupt current treatment protocols but could enable combination therapies with drugs that increase NQO1 expression, enhancing the efficacy of IB-DNQ. From a clinical management perspective, the use of minimally invasive biopsies in DIPG could provide valuable insights into key mutations or protein overexpression patterns, leading to more personalized treatment approaches. For instance, patients with PPM1D truncations associated with NQO1 upregulation could potentially benefit from the administration of IB-DNQ or similar agents. Importantly, IB-DNQ has shown low toxicity in animal models, and given the urgent need for new therapeutic strategies in DIPG, these findings make IB-DNQ a promising candidate for further research.
Conclusion
In conclusion, this study identifies PPM1D truncation as a key driver of NQO1 overexpression in DIPG and demonstrates the efficacy of the NQO1-bioactivatable compound IB-DNQ in targeting these cells. While further investigation is needed to clarify the molecular interactions involved, the potential of combining IB-DNQ with existing therapies offers a promising approach for treating patients with PPM1D mutations. Given the low toxicity of IB-DNQ in animal models and the critical need for new DIPG therapies, these findings provide a strong foundation for future preclinical and clinical studies.
Acknowledgments
Gratitude is extended to Dr. Andres Morales La Madrid, Dr. Angel Montero Carcaboso and Dr. Jaume Mora from the Pediatric Cancer Center Barcelona (PCCB) at Hospital Sant Joan de Déu, Barcelona for their significant support. I thank Dr. Michelle Monje from Stanford Medicine for providing SU-DIPG-VI cell line and SU-DIPG-35 frozen tissue and matched normal cortex. I warmly thank Dr. Pratiti Bandopadhayay, Dr. Eric Morin, and Dr. Timothy Phoenix from the Boston Dana-Farber Cancer Institute for providing data regarding IUE mouse models. I acknowledge Dr. Chris Jones from the Institute of Cancer Research, London, for sharing somatic mutations data information regarding DIPG cell lines. I thank Dr. Myung Ryul Lee and Dr. Paul J. Hergenrother for providing IB-DNQ compound and UZH’s DMG Research Center, University Children’s Hospital Zurich for providing tumor samples from patients, especially Dr. Sulayman Mourabit, Dr. Javad Nazarian, and Dr. Denise Morinigo from the Children’s National Hospital, Washington D.C. I also thank CERCA Program/ Government of Catalonia for their institutional support.
Grant Support
M. Janin received support from the Spanish Ministry of Science, Innovation and Universities for his Sara Borrell postdoctoral fellowship. This work has been supported by EPIC-XS, project number 823839, funded by the Horizon 2020 programme of the European Union.
Data Availability
All data generated for this study are presented in the article.
Conflicts of Interest
The author does not report conflicts of interest.
Authors Contributions
M. Janin was responsible for the conception and design of the study, as well as the development of the methodology. M. Janin also handled the acquisition of data, including managing patient data and providing necessary facilities, and conducted the analysis and interpretation of data through statistical and computational methods. Additionally, M. Janin contributed to the writing, review, and revision of the manuscript, provided administrative, technical, and material support, and supervised the overall study.
References
- Rashed WM, Maher E, Adel M, Saber O, Zaghloul MS. Pediatric diffuse intrinsic pontine glioma: Where do we stand?. Cancer Metastasis Rev. 2019;38(4):759-770.
[Crossref] [Google Scholar] [PubMed]
- Vanan MI, Eisenstat DD. DIPG in children–what can we learn from the past?. Front Oncol. 2015;5:237.
[Crossref] [Google Scholar] [PubMed]
- Jones C, Karajannis MA, Jones DT, Kieran MW, Monje M, Baker SJ, et al. Pediatric high-grade glioma: Biologically and clinically in need of new thinking. Neuro Oncol. 2017;19(2):153-161.
[Crossref] [Google Scholar] [PubMed]
- Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7(1):11185.
[Crossref] [Google Scholar] [PubMed]
- El-Khouly FE, Veldhuijzen van Zanten SE, Santa-Maria Lopez V, Hendrikse NH, Kaspers GJ, Loizos G, et al. Diagnostics and treatment of diffuse intrinsic pontine glioma: Where do we stand?. J Neurooncol. 2019;145:177-184.
[Crossref] [Google Scholar] [PubMed]
- Janin M, Ortiz-Barahona V, de Moura MC, Martínez-Cardús A, Llinàs-Arias P, Soler M, et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053-1074.
[Crossref] [Google Scholar] [PubMed]
- Lundberg AP, Boudreau MW, Selting KA, Chatkewitz LE, Samuelson J, Francis JM, et al. Utilizing feline oral squamous cell carcinoma patients to develop NQO1-targeted therapy. Neoplasia. 2021;23(8):811-822.
[Crossref] [Google Scholar] [PubMed]
- Lundberg AP, Francis JM, Pajak M, Parkinson EI, Wycislo KL, Rosol TJ, et al. Pharmacokinetics and derivation of an anticancer dosing regimen for the novel anti-cancer agent isobutyl-deoxynyboquinone (IB-DNQ), a NQO1 bioactivatable molecule, in the domestic felid species. Invest New Drugs. 2017;35(2):134-144.
[Crossref] [Google Scholar] [PubMed]
- Martínez-Reyes I, Chandel NS. Cancer metabolism: Looking forward. Nat Rev Cancer. 2021;21(10):669-680.
[Crossref] [Google Scholar] [PubMed]
- Preethi S, Arthiga K, Patil AB, Spandana A, Jain V. Review on NAD (P) H dehydrogenase quinone 1 (NQO1) pathway. Mol Biol Rep. 2022;49(9):8907-8924.
[Crossref] [Google Scholar] [PubMed]
- Glorieux C, Sandoval JM, Dejeans N, Ameye G, Poirel HA, Verrax J, et al. Overexpression of NAD (P) H: Quinone oxidoreductase 1 (NQO1) and genomic gain of the NQO1 locus modulates breast cancer cell sensitivity to quinones. Life Sci. 2016;145:57-65.
[Crossref] [Google Scholar] [PubMed]
- Ji M, Jin A, Sun J, Cui X, Yang Y, Chen L, et al. Clinicopathological implications of NQO1 overexpression in the prognosis of pancreatic adenocarcinoma. Oncol Lett. 2017;13(5):2996-3002.
[Crossref] [Google Scholar] [PubMed]
- Siegel D, Franklin WA, Ross D. Immunohistochemical detection of NAD (P) H: Quinone oxidoreductase in human lung and lung tumors. Clin Cancer Res. 1998;4(9):2065-2070.
[Google Scholar] [PubMed]
- Siegel D, Ross D. Immunodetection of NAD (P) H: Quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic Biol Med. 2000;29(3-4):246-253.
[Crossref] [Google Scholar] [PubMed]
- Buranrat B, Chau-in S, Prawan A, Puapairoj A, Zeekpudsa P, Kukongviriyapan V. NQO1 expression correlates with cholangiocarcinoma prognosis. Asian Pac J Cancer Prev. 2012;13(Suppl):131-136.
[Google Scholar] [PubMed]
- Cui X, Li L, Yan G, Meng K, Lin Z, Nan Y, et al. High expression of NQO1 is associated with poor prognosis in serous ovarian carcinoma. BMC Cancer. 2015;15:244.
[Crossref] [Google Scholar] [PubMed]
- Li Z, Zhang Y, Jin T, Men J, Lin Z, Qi P, et al. NQO1 protein expression predicts poor prognosis of non-small cell lung cancers. BMC Cancer. 2015;15:207.
[Crossref] [Google Scholar] [PubMed]
- Krajka-Kuźniak V, Paluszczak J, Baer-Dubowska W. The Nrf2-ARE signaling pathway: An update on its regulation and possible role in cancer prevention and treatment. Pharmacol Rep. 2017;69(3):393-402.
[Crossref] [Google Scholar] [PubMed]
- Carvalho DM, Temelso S, Mackay A, Pemberton HN, Rogers R, Kessler K, et al. Drug screening linked to molecular profiling identifies novel dependencies in patient-derived primary cultures of paediatric high grade glioma and DIPG. bioRxiv. 2020;2020-12.
- Zhang L, Chen LH, Wan H, Yang R, Wang Z, Feng J, et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet. 2014;46(7):726-730.
[Crossref] [Google Scholar] [PubMed]
- Kahn JD, Miller PG, Silver AJ, Sellar RS, Bhatt S, Gibson C, et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood. 2018;132(11):1095-1105.
[Crossref] [Google Scholar] [PubMed]
- Kleiblova P, Shaltiel IA, Benada J, Evčík J, Pecháčková S, Pohlreich P, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013;201(4):511-521.
[Crossref] [Google Scholar] [PubMed]
- Dudgeon C, Shreeram S, Tanoue K, Mazur SJ, Sayadi A, Robinson RC, et al. Genetic variants and mutations of PPM1D control the response to DNA damage. Cell Cycle. 2013;12(16):2656-2664.
[Crossref] [Google Scholar] [PubMed]
- Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19(10):1162-1174.
[Crossref] [Google Scholar] [PubMed]
- Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99(5):3099-3104.
[Crossref] [Google Scholar] [PubMed]
- Patiño-Morales CC, Soto-Reyes E, Arechaga-Ocampo E, Ortiz-Sánchez E, Antonio-Véjar V, Pedraza-Chaverri J, et al. Curcumin stabilizes p53 by interaction with NAD (P) H: Quinone oxidoreductase 1 in tumor-derived cell lines. Redox Biol. 2020;28:101320.
[Crossref] [Google Scholar] [PubMed]
- Akamandisa MP, Nie K, Nahta R, Hambardzumyan D, Castellino RC. Inhibition of mutant PPM1D enhances DNA damage response and growth suppressive effects of ionizing radiation in diffuse intrinsic pontine glioma. Neuro Oncol. 2019;21(6):786-799.
[Crossref] [Google Scholar] [PubMed]
- Khadka P, Reitman ZJ, Lu S, Buchan G, Gionet G, Dubois F, et al. PPM1D mutations are oncogenic drivers of de novo diffuse midline glioma formation. Nat Commun. 2022;13(1):604.
[Crossref] [Google Scholar] [PubMed]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12(4):342-354.
[Crossref] [Google Scholar] [PubMed]
- Gu L, Zhang H, He J, Li J, Huang M, Zhou M. MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene. 2012;31(11):1342-1353.
[Crossref] [Google Scholar] [PubMed]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32(2):180-189.
[Crossref] [Google Scholar] [PubMed]
- Huang X, Dong Y, Bey EA, Kilgore JA, Bair JS, Li LS, et al. An NQO1 substrate with potent antitumor activity that selectively kills by PARP1-induced programmed necrosis. Cancer Res. 2012;72(12):3038-3047.
[Crossref] [Google Scholar] [PubMed]
- Parkinson EI, Hergenrother PJ. Deoxynyboquinones as NQO1-activated cancer therapeutics. Acc Chem Res. 2015;48(10):2715-2723.
[Crossref] [Google Scholar] [PubMed]
- Bair JS, Palchaudhuri R, Hergenrother PJ. Chemistry and biology of deoxynyboquinone, a potent inducer of cancer cell death. J Am Chem Soc. 2010;132(15):5469-5478.
[Crossref] [Google Scholar] [PubMed]
- Yang Y, Zhou X, Xu M, Piao J, Zhang Y, Lin Z, et al. β-lapachone suppresses tumour progression by inhibiting epithelial-to-mesenchymal transition in NQO1-positive breast cancers. Sci Rep. 2017;7(1):2681.
[Crossref] [Google Scholar] [PubMed]
- Warren KE. Beyond the blood brain barrier: The importance of Central Nervous System (CNS) pharmacokinetics for the treatment of CNS tumors, including diffuse intrinsic pontine glioma. Front Oncol. 2018;8:239.
[Crossref] [Google Scholar] [PubMed]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931-947.
[Crossref] [Google Scholar] [PubMed]
- Oh ET, Park HJ. Implications of NQO1 in cancer therapy. BMB Rep. 2015;48(11):609-617.
[Crossref] [Google Scholar] [PubMed]
- Terai K, Dong GZ, Oh ET, Park MT, Gu Y, Song CW, et al. Cisplatin enhances the anticancer effect of β-lapachone by upregulating NQO1. Anticancer Drugs. 2009;20(10):901-909.
[Crossref] [Google Scholar] [PubMed]
- Zeekpudsa P, Kukongviriyapan V, Senggunprai L, Sripa B, Prawan A. Suppression of NAD (P) H-quinone oxidoreductase 1 enhanced the susceptibility of cholangiocarcinoma cells to chemotherapeutic agents. J Exp Clin Cancer Res. 2014;33:11.
[Crossref] [Google Scholar] [PubMed]
- Choi EK, Terai K, Ji IM, Kook YH, Park KH, Oh ET, et al. Upregulation of NAD (P) H: Quinone oxidoreductase by radiation potentiates the effect of bioreductive β-lapachone on cancer cells. Neoplasia. 2007;9(8):634-642.
[Crossref] [Google Scholar] [PubMed]
- Claerhout S, Lim JY, Choi W, Park YY, Kim K, Kim SB, et al. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PloS One. 2011;6(9):e24662.
[Crossref] [Google Scholar] [PubMed]
Citation: Janin M (2024). PPM1D Truncation-associated Overexpression of the Stress-related Protein NQO1 Confers Sensitivity to the Bioactivatable Drug IB-DNQ in Diffuse Intrinsic Pontine Glioma. J Clin Exp Pharmacol. 14:440.
Copyright: © 2024 Janin M. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.