Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Proquest Summons
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 15, Issue 2
Incidence of CD36 Antibody Induced Alloimmunisation and CD36 Deficiency Distribution among Different Populations: A Systematic Review and Meta-Analysis
Yutian Wang and Denise E. Jackson*Received: 22-Jan-2024, Manuscript No. JBDT-24-24630; Editor assigned: 24-Jan-2024, Pre QC No. JBDT-24-24630 (PQ); Reviewed: 14-Feb-2024, QC No. JBDT-24-24630; Revised: 21-Feb-2024, Manuscript No. JBDT-24-24630 (R); Published: 28-Feb-2024, DOI: 10.4172/2155-9864.24.15.578
Abstract
Alloimmunisation, such as Platelet Transfusion Refractoriness (PTR), is an immune response to foreign compounds upon host exposure to foreign antigens. Alloimmunisation can cause life-threatening effects in patients. Recent studies have reported alloimmunisation from CD36 deficiency, and the risk of alloimmunisation produced by anti- CD36 antibodies. This study aimed to determine the risk of anti-CD36 antibody-induced alloimmunisation in global populations. This systematic review used the Embase, PubMed, Scopus, and Web of Science as databases to search for relevant articles from 1995 to 2023. The meta-analysis used OpenMeta Analyst software to perform the one-arm analysis with binary random effect and maximum likelihood methods using arcsine transformed proportion.
Alloimmunisation caused by CD36 antibody, with an overall rate of 0.83% (P-value<0.001; I2=79.71; 95% C.I. [0.002, 0.027]). CD36 gene mutation rate in the CD36 deficiency population (rate=65.22%, P-value<0.001, I2=75.52, 95% C.I. [0.554, 0.836]). CD36 deficiency rate in different ethnicities included Asians with 2.90% (P-value<0.001, I2=85.49, 95% C.I. [0.020, 0.033]), 4.59% in African (P-value<0.001, I2=36.31, 95% C.I. [0.032, 0.060]) and 0.61% in Caucasian (P-value=0.013, I2=92.23, 95% C.I. [0.000, 0.020]). The rate of deficiency among different areas of Asia: 3.51% for North-East Asia (P-value<0.001, I2=63.07, 95% C.I. [0.022, 0.059]) and 3.62% for South-East Asia (P-value<0.001, I2=83.82, 95% C.I. [0.025, 0.039]). This study concluded that CD36 deficient populations with gene mutations are at risk of anti-CD36 alloimmunisation and Fetal and Neonatal Alloimmune Thrombocytopenia (FNAIT).
Keywords
Platelet transfusion refractoriness; PTR; Fetal and Neonatal Alloimmune Thrombocytopenia (FNAIT); Alloimmunisation; Anti-CD36; CD36 antibody; CD36 deficiency; CD36 gene mutation
Introduction
Alloimmunisation occurs when a patient is exposed to a foreign antigen via transfusion or pregnancy [1]. Platelet-mediated alloimmunisation, such as platelet transfusion refractoriness or Fetal and Neonatal Alloimmune Thrombocytopenia (FNAIT), can cause severe outcomes such as long-term hospitalisation, low efficacy of treatment, and high risks of bleeding [2]. Duke first reported platelet transfusion in a case of maintaining haemostasis in a 20-year-old patient to confront the issue of bleeding [3]. Platelet transfusion can be used for thrombocytopenic patients with acutely low platelet survival lifespan or low platelet counts to avoid an unbalanced haemostasis state after some therapy such as chemotherapy [4]. Europe produces roughly 2.9 million platelet products each year [5]. Platelet transfusion refractoriness means platelet counts cannot reach the expected level after platelet transfusion. There are three methods to estimate the outcomes of platelet transfusions. One method is to measure the Corrected Count Increment (CCI). If the patient’s CCI remains less than 5-10 × 109/L after one hour of platelet transfusion, platelet transfusion refractoriness can be defined [6]. In addition, a Percentage Platelet Recovery (PPR) lower than 20% at 20-40 hours or 30% at one hour indicates platelet refractoriness [4]. Another indicator is post-transfusion Platelet Increment (PI). Platelet refractoriness is determined if PI is less than 10 × 109/L at 24 h [6]. The causes of alloimmunisation can be divided into immune and non- immune groups. Most cases are associated with non-immune factors such as fever, drugs and graft-versus-host disease [2]. Approximately 10%-20% of cases are caused by immune factors, such as ABO incompatibility, Human Leukocyte Antigens (HLA) alloimmunisation by antibodies against human leukocyte antigen class I, and antibodies against human platelet antigens. Recently, an antibody against CD36 on platelets, which induces alloimmunisation or platelet transfusion refractoriness, has gained interest.
Characteristics of CD36 antibody and associated clinical outcomes
CD36 is a term for Glycoprotein IV (GPIV), Fatty Acid Translocase (FAT), and Glycoprotein IIIb (GPIIIB) and is expressed on all surfaces of differentiated cells. CD36 is an 88-kDa integral membrane protein with multiple ligands [7]. It is a class B scavenger receptor family member with one extracellular loop in two transmembrane domains [7]. CD36 genes located on chromosome 7q11.2 produce this glycoprotein [8]. Fifteen exons are involved in CD36 gene [9]. The deficiency of CD36 in humans was first introduced by a case study in which platelet transfusion refractoriness was induced in a patient by multiple transfusions of platelets [10]. The CD36 deficiency can be classified as type I and type II. Type I indicates that both platelets and monocytes lack expression, whereas in type II, only CD36 surface expression on platelets is repressed [11]. Most cases of CD36 deficiency are associated with the African and Asian populations. This human gene mutation has been linked to many different diseases. Dr. Koji Miyaoka’s team investigated the linkage between insulin resistance and CD36 genetic deficiency, as CD36 is a major receptor on macrophages and is responsible for the metabolism of long-chain fatty acids [12]. Furthermore, CD36 is also an essential target for Plasmodium falciparum, which causes the most severe malaria. Hence, CD36 deficiency in the African region might result from selection pressure [13]. Only individuals lacking CD36expression can establish CD36 isoimmunisation, although they are healthy. When the patient is exposed to CD36 antigen on platelets through platelet transfusion, the immune system can produce CD36 antibodies and cause alloimmunisation. Most refractory patients will develop alloimmunisation by the CD36 type I deficiency phenotype [14].
Methods to determine CD36 deficiency
Different institutions use different methods to determine CD36 deficiencies. However, most methods followed a similar strategy: testing expression in platelets and mutant genes. For example, the technology used by the Suzhou Blood Centre to phenotype CD36 on human platelets is sandwich Enzyme-Linked Immunosorbent Assay (ELISA). ELISA detects an antibody that targets CD36 antigen on platelets by binding to an enzyme. The enzyme converted colourless products to colourful products, indicating the presence of CD36 [15]. Flow Cytometry (FC) is another technology used to confirm this deficiency [9]. The principle of FC is to distinguish the CD36 antigen by light emission from fluorescently labelled antibodies of different sizes and intensities [16]. Moreover, Polymerase Chain Reaction Sequence-Based Typing (PCR-SBT) is used to analyse gene mutation of CD36 deficiency individuals by accurately examining the nucleotide sequence of the alleles [9,17].
Platelet transfusion is a widely used practice in Australian hospitals to maintain haemostasis in patients with thrombocytopenia. Alloimmunisation, such as platelet transfusion refractoriness, can result in life-threatening outcomes for patients. Therefore, it is essential to avoid all factors that can cause alloimmunisation. Patients without CD36 expression on platelets can also develop anti-CD36 antibodies during platelet transfusion or pregnancy. This highlights the importance of evaluating the incidence. However, owing to the high cost of CD36 deficiency testing and the urgency of platelet transfusion, it is impossible to test for CD36 deficiency in each patient who needs platelet transfusion or pregnancy. Australia is a multicultural country with immigrants of various ethnicities. Patients might have distinct backgrounds, so the gene may vary and lead to varied expressions of CD36. By testing the CD36 deficiency distribution in blood donors, the relationship between ethnicity and CD36 deficiency can be determined. Hence, when patients have a background of high CD36 deficiency risk induced by ethnicity, possible alloimmunisation should be considered, and a better outcome of platelet transfusion can be achieved. The primary aim of this review and the research question are: What is the incidence of alloimmunisation caused by antibodies against CD36 on platelets? To achieve this aim, the study will analyse several cases induced by CD36 antibodies in all immunisation cases. As the alloimmunisation caused by CD36 antibody is associated with CD36 deficiency, the secondary research question is: What are the differences in CD36 deficiency distribution among different ethnicities? This study examined the frequency of CD36 deficiency in platelet donors in different countries and areas. Based on these research questions, the hypothesis is that antibodies against CD36 on platelets can cause alloimmunisation, and Asian and African populations are more likely to have CD36 deficiency.
Materials and Methods
Study design
The Preferred Reporting Item for Systematic Review and Meta-analysis (PRISMA) procedure was applied in this systematic review to collect relevant papers indicating the incidence of CD36 antibody-induced alloimmunisation and CD36 deficiency distribution among different populations [18]. Articles included in this systematic review were also assessed for quality using the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) checklist [19].
Search strategy protocol
The Embase, PubMed, Scopus, and Web of Science databases were used to search for relevant articles, and the years ranged from 1995 to 2023. The search dates were not limited because of the low number of relevant articles. “Alloimmunisation” and “CD36,” “platelet transfusion refractoriness” and “CD36” or “CD36 deficiency,” and “platelet refractoriness” and “CD36” were used as crucial search terms in the search process. The word “Alloimmunisation” is spelled as “Alloimmunization” in American English, and “CD36 deficiency” could also be interpreted as “platelet glycoprotein deficiency.” A manual search was performed by reviewing the bibliography when other studies were mentioned in the selected literature and identified by the RMIT library system.
Inclusion and exclusion criteria
The exclusion criteria followed the PRISMA flowchart [18]. All papers from the database searches were imported into EndNote for duplicate removal. To validate the eligibility of articles from the database searches, all articles were first screened based on the title and abstract. Articles that did not mention the alloimmunisation rate caused by CD36 or CD36 deficiency investigation in subjects in the abstract were excluded. Further screening was done to ensure the eligibility of literature based on this criterion; excluded articles with no relevant data provided, no article found or not English articles, not mentioned “alloimmunisation” or “CD36,” and only case studies in articles. After the exclusion, the eligible articles were included based on the following criteria: a. Articles had an alloimmunisation rate caused by anti-CD36 antibodies among the population instead of the case study; b. Articles included gene mutation rate in the CD36 deficiency population, c. The CD36 deficiency rate among different ethnicities of the population.
Data selection criteria
The selected articles obtained all the data used in this systematic review. The first author, publication year, study design, country of publication, study period, sample size, and measured parameters were included in this review. The sample size was determined at an alloimmunisation rate due to CD36 antibody or CD36 deficiency rate among specific ethnicities in the study. The parameters used in the analysis included the number of alloimmunisation caused by anti-CD36, alloimmunisation type (e.g., platelet transfusion refractoriness, neonatal alloimmune thrombocytopenia), number of CD36 deficiencies, ethnicity of CD36 deficiency population, and rate of CD36 gene mutation. Some articles have reported the rates of CD36 deficiency among different provinces in China. Only the northernmost and southernmost data were included, as the remaining areas were not representative.
Statistical analysis
This review aimed to investigate the correlation between CD36 antibodies and alloimmunisation; which area of the population is most likely to have CD36 deficiency? A meta-analysis needs to be conducted in this review. Open Meta-Analyst software from Brown University was used to perform the meta-analysis [20]. The meta-analysis used arcsine transformed proportion, binary random effects, and maximum likelihood methods to perform a one-arm analysis for each parameter. One-arm analysis was performed to determine the risk of alloimmunisation caused by CD36 antibody; the rate of CD36 deficiency among Asian, African, and Caucasian populations; the rate of CD36 deficiency among North-East Asia and South-East Asia; and the rate of gene mutation in CD36 deficiency individuals. Forest plots showed the results of one-arm analysis with an overall P-value of less than 0.05, which was deemed statistically significant; 95% confidence intervals (95% CI), I2 with P-value to indicate heterogeneity.
Results
Study screened
The article selection process used in this review follows the Prisma flow chart shown in Figure 1. From these four databases, 3348 articles were obtained based on advanced keyword searches. After importing all the articles into EndNote, 920 duplicates were identified and excluded. The remaining studies then underwent screening of titles and abstracts, and another 127 articles were removed from the potential list. Only 2301 studies were evaluated for eligibility. By assessing the content of each paper, 15 articles were excluded due to lack of relevant data support, 36 papers were removed as only case studies were involved, another 36 studies were excluded based on not being appropriate with the research hypothesis, and 24 studies were excluded because of the lack of articles provided or languages. Finally, 12 articles passed eligibility screening, and another four studies were also included by manual searching; the total number of studies included in the meta-analysis was 16.
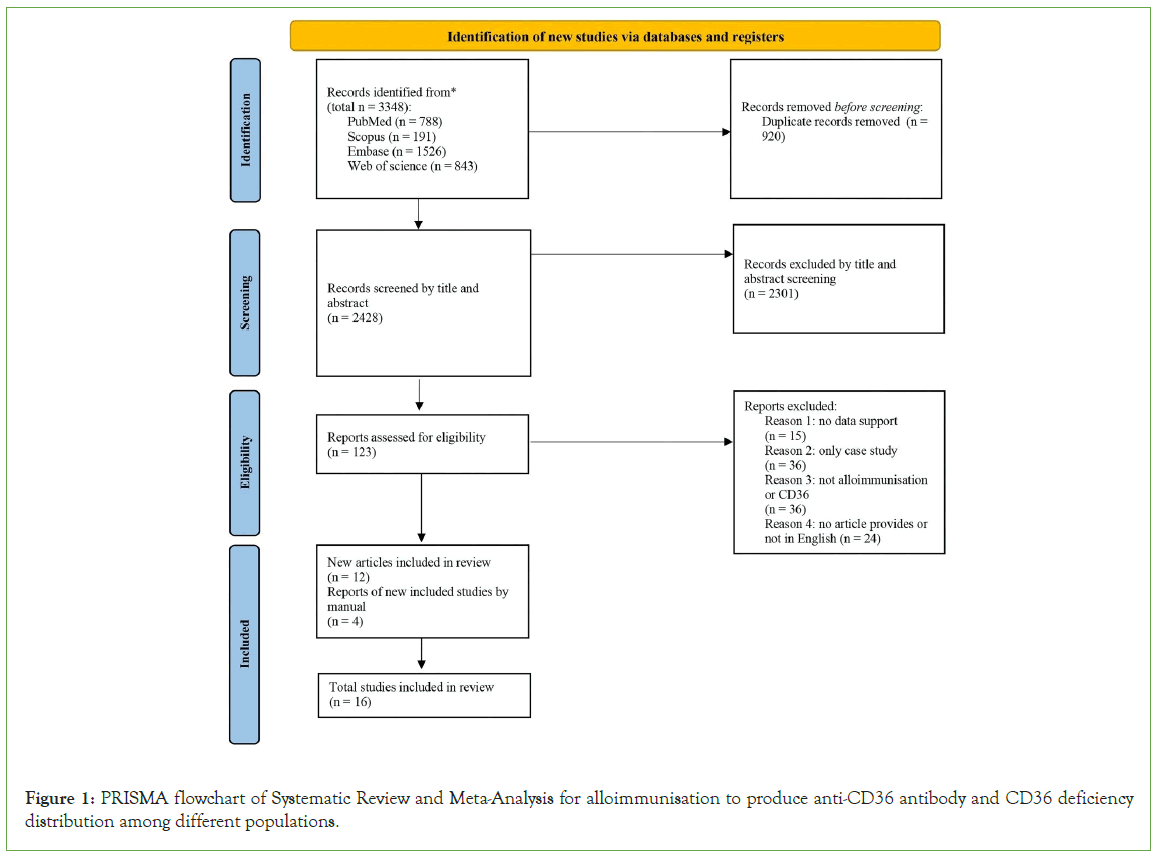
Figure 1: PRISMA flowchart of Systematic Review and Meta-Analysis for alloimmunisation to produce anti-CD36 antibody and CD36 deficiency distribution among different populations.
Study characteristics
The analysis of alloimmunisation by CD36 antibody and CD36 deficiency distribution and summarises the characteristics of each eligible article (Table 1). Most studies use a cohort study design, while others do not mention the study design. Almost all the included articles were published before the 21st century. These studies were performed in eight different countries; and each article listed the specific ethnicity of the study subjects. The total sample sizes varied from 55 to 7409 subjects, while the number of alloimmunisation ranged from 1 to 15, and the number of CD 36 deficiencies was between 0 and 259 individuals. However, only four articles included alloimmunisation cases by anti-CD36, whereas other studies measured CD36 deficiency rates and gene mutation rates among CD36 deficiency individuals.
| Study | Study design | Country | Study period | Sample size | Parameter measured |
|---|---|---|---|---|---|
| Ohto et al; 2004 [34] | Cohort study | Japan | 1990-2002 | 2/223 | No., type of alloimmunisation |
| Choi et al; 2014 [21] | / | Korea | 2014-2014 | 5/180 | No., area of CD36 deficiency |
| Curtis et al; 1996 [22] | / | USA | / | 6/530 | No., area of CD36 deficiency |
| Flesch et al; 2021 [29] | / | Arabia | 2016-2018 | 1/55a 35/985b |
No., type of alloimmunisation, No., area of CD36 deficiency, gene mutation rate |
| He et al; 2023 [9] | / | China | 2020-2021 | 36/1691 | No., area of CD36 deficiency, gene mutation rate |
| Lee et al; 1999 [35] | Cohort study | France | / | 4/1885a 34/861b |
No., type of alloimmunisation, No., area of CD36 deficiency |
| Li et al; 2022 [23] | Cohort study | China | 2007-2019 | 209/5081 | No., area of CD36 deficiency |
| Liu et al; 2020 [32] | Cohort study | China | 2018-2020 | 85/5315 | No., area of CD36 deficiency, gene mutation rate |
| Lyu et al; 2023 [36] | Cohort study | China | 2020-2021 | 7/418 | No., area of CD36 deficiency, gene mutation rate |
| Lo et al; 2016 [37] | / | China | / | 10/640 | No., area of CD36 deficiency, gene mutation rate |
| Ma et al; 2019 [38] | Retrospective | China | 2017-2017 | 13/612 | No., area of CD36 deficiency |
| Masuda et al; 2015 [39] | / | Japan | / | 11/135 | No., area of CD36 deficiency, gene mutation rate |
| Phuangtham et al; 2020 [31] | / | Thailand | / | 15/700 | No., area of CD36 deficiency, gene mutation rate |
| Wu et al; 2017 [26] | / | China | 2008-2015 | 15/478a 254/7409b |
No., type of alloimmunisation, No., area of CD36 deficiency |
| Xia et al; 2016 [30] | / | China | / | 36/1391 | No., area of CD36 deficiency, gene mutation rate |
| Yamamoto et al; 1994 [40] | / | Japan | / | 16/354 | No., area of CD36 deficiency |
Note: a: Alloimmunisation cases caused by anti-CD36 antibody; b: CD36 deficiency rate among all test population.
Table 1: Characteristics of eligible studies for analyzing alloimmunisation by anti-CD36 antibody and CD36 deficiency distribution.
All the data included in the meta-analysis are listed in Table 2. The numbers of alloimmunisation are shown in four studies, and the specific type of alloimmunisation caused by anti-CD36 is also mentioned. The remaining studies analysed the correlation between CD36 deficiency and ethnicity, especially in Asia. In addition, the gene mutation rate among patients with CD36 deficiency was collected from eight studies to conduct the meta-analysis.
| Study | No. of alloimmunisation | Type of casesc | No. of CD36 deficiency | Ethnicity of CD36 deficiency cases | Gene mutation rate |
|---|---|---|---|---|---|
| Ohto et al; 2004 [34] | 2/223 | FMAIT | / | / | / |
| Choi et al; 2014 [21] | / | / | 5/180 | Korea | / |
| Curtis et al; 1996 [22] | / | / | 6/250 0/280 |
African American Caucasian |
/ |
| Flesch et al; 2021 [29] | Jan-55 | NAIT | 31/946 4/39 |
Caucasianc Africand |
34/35 |
| He et al; 2023 [9] | / | / | 36/1691 | China (Suzhou) | 13/27 |
| Lee et al; 1999 [35] | 4/1885 | PTP(2), PTR(1), NAIT(1) | 1/301 33/560 |
European African, Afro-Caribbeans |
/ |
| Li et al; 2022 [23] | / | / | 209/5081 | China (Nanning) | / |
| Liu et al; 2020 [32] | / | / | 85/5315 | China | 31/61 |
| Lyu et al; 2023 [36] | / | / | 7/418 | China (Kunming) | 6/7 |
| Lo et al; 2016 [37] | / | / | 10/640 | China (Taiwan) | 8/10 |
| Ma et al; 2019 [38] | / | / | 13/612 259/7549 4/4372 40/958 |
Northern China Southern China Caucasian African |
/ |
| Masuda et al; 2015 [39] | / | / | 11/135 | Japan | 8/11 |
| Phuangtham et al; 2020 [31] | / | / | 15/700 | Thailand | 9/15 |
| Wu et al; 2017 [26] | 15/478 | NAIT(7) PTR(7) Donor with Ab(1) |
254/7409 190/4621 |
China Guangxi |
/ |
| Xia et al; 2016 [30] | / | / | 36/1391 | South-East Asia | 11/18 |
| Yamamoto et al; 1994 [40] | / | / | 16/354 | Japan | / |
Note: c: Syria, Arabian Peninsula, Northern Africa belong to Caucasian population; d: Sub-Saharan Africa belongs to African population.
Table 2: Data of eligible studies included in meta-analysis reporting rate of alloimmunisation by anti-CD36 antibody and CD36 deficiency distribution.
Quality assessment of the included studies
The methodological quality of the selected studies was evaluated using the STROBE Checklist (Table 3). The STROBE checklist helped to avoid low-quality articles with unsuitable data to be included in the review [19]. The titles and abstracts of all the articles provided a critical summary of the content, and all the selected articles had high-quality introductions that provided the background or rationale of the studies and stated objectives or hypotheses. Most studies have characterised study methods, participant selection criteria, statistical methods, and potential bias on the methods part. However, two studies did not introduce the statistical methods, whereas another article failed to address this bias [21-23]. Furthermore, the above three articles did not provide an interpretation of the limitations in the discussion section, while the rest addressed them well. Nevertheless, these three articles exhibited good enough overall quality. Therefore, all the studies met the quality criteria and were eligible for the meta-analysis.
| Title and abstract | Introduction | Methods | Results | Discussion | ||||
|---|---|---|---|---|---|---|---|---|
| Clear title and abstract with study design indicated | Explain scientific background | Detailed study methods given | Eligibility criteria for participant selection shown | Describes statistical methods | Describes any efforts to address potential bias | Give characteristic of study participants | Summarise key results and discusses limitations | |
| Ohto et al; 2004 [34] | Y | Y | Y | Y | Y | Y | Y | Y |
| Choi et al; 2014 [21] | Y | Y | Y | Y | N | Y | Y | Ne |
| Curtis et al; 1996 [22] | Y | Y | Y | Y | N | Y | Y | Ne |
| Flesch et al; 2021 [29] | Y | Y | Y | Y | Y | Y | Y | Y |
| He et al; 2023 [9] | Y | Y | Y | Y | Y | Y | Y | Y |
| Lee et al; 1999 [35] | Y | Y | Y | Y | Y | Y | Y | Y |
| Li et al; 2022 [23] | Y | Y | Y | Y | Y | N | Y | Ne |
| Liu et al; 2020 [32] | Y | Y | Y | Y | Y | Y | Y | Y |
| Lyu et al; 2023 [36] | Y | Y | Y | Y | Y | Y | Y | Y |
| Lo et al; 2016 [37] | Y | Y | Y | Y | Y | Y | Y | Y |
| Ma et al; 2019 [38] | Y | Y | Y | Y | Y | Y | Y | Y |
| Masuda et al; 2015 [39] | Y | Y | Y | Y | Y | Y | Y | Y |
| Phuangtham et al; 2020 [31] | Y | Y | Y | Y | Y | Y | Y | Y |
| Wu et al; 2017 [26] | Y | Y | Y | Y | Y | Y | Y | Y |
| Xia et al; 2016 [30] | Y | Y | Y | Y | Y | Y | Y | Y |
| Yamamoto et al; 1994 [40] | Y | Y | Y | Y | Y | Y | Y | Y |
Note: Y-criteria fulfilled; N-criteria not fulfilled; e-No limitations discussed.
Table 3: Evaluation of methodological quality of included studies according to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) checklist.
Meta-analysis on the risk of alloimmunisation caused by CD36 deficiency and gene mutation rate in CD36 deficiency population
mThe data of alloimmunisation cases caused by CD36 deficiency from Table 2, were used to generate a meta-analysis to investigate the incidence of CD36 antibody-induced alloimmunisation. The forest plot was constructed using arcsine transformed proportions and binary random effects (Figure 2). The blue diamond shape indicates the overall alloimmunisation rate in the Figure 2. The P-value of the overall one-arm analysis was less than 0.001, which was significant. The incidence of causing alloimmunisation by anti-CD36 ranged from 0.21% to 3.14% in each study, and the total incidence calculated by adding all the alloimmunisation cases from four studies was 0.83%. The high heterogeneity of this forest plot, indicated by a P-value of less than 0.001 and I2 of 79.71, determined the diversity of the selected data. However, from the low 95% C.I. value (0.002, 0.027), the incidence of causing alloimmunisation by anti-CD36 was relatively low.

Figure 2: A one-arm proportion forest plot was analysed under the random effects model with the maximum likelihood method for alloimmunisation caused by anti-CD36 antibodies and CD36 gene mutation rates (A) Alloimmunisation rate that produces anti-CD36 antibody; (B) CD36 gene mutation rate among CD36 deficiency population.
Figure 2, shows the one-arm forest plot of gene mutation rates among the CD36 deficiency population from eight studies. The total incidence of CD36 gene mutation was 65.22%, and the overall P-value of the forest plot was significant (P-value<0.001). The dataset heterogeneity was high, as I2 was 75.52 (P-value<0.001). 95% C.I. was 0.554, 0.836. The incidence of CD36 gene mutations ranged from 50.82% to 97.14%.
Meta-analysis on the rate of CD36 deficiency among different ethnicities
The difference in CD36 deficiency among Asians, Africans, and Caucasians was determined using three forest plots in Figure 3. Figure 3A, shows the one-arm analysis with arcsine transformed proportion determining the incidence of CD36 deficiency among the Asian population from 13 articles. The overall P-value was less than 0.001, the P-value of heterogeneity was less than 0.001, and the I2 value was 85.49. This indicated that the overall data were significant, and high heterogeneity showed a divergence of data (95% C.I. 0.020, 0.033). The total CD36 deficiency ratio in Asia calculated in 13 studies was 2.90%. CD36 deficiency among the African population was analysed by arcsine proportion, as shown in Figure 3B. The overall P-value was significant (<0.001), the I2 value (36.31) was relatively low, and its P-value was equal to 0.05, which means it was statistically non-significant; hence, the data were close within these studies (95% C.I. 0.032, 0.060). The total incidence rate in the African population was 4.59%. Four studies were used to construct forest plots in Figure 3C, with a P-value of 0.013; the overall P-value was significant. I2 was 92.23, while its P-value was less than 0.001. These results indicate a high difference between the data from each article. The overall ratio of CD36 deficiency among Caucasian patients was 0.61% (95% C.I. 0.000, 0.020).
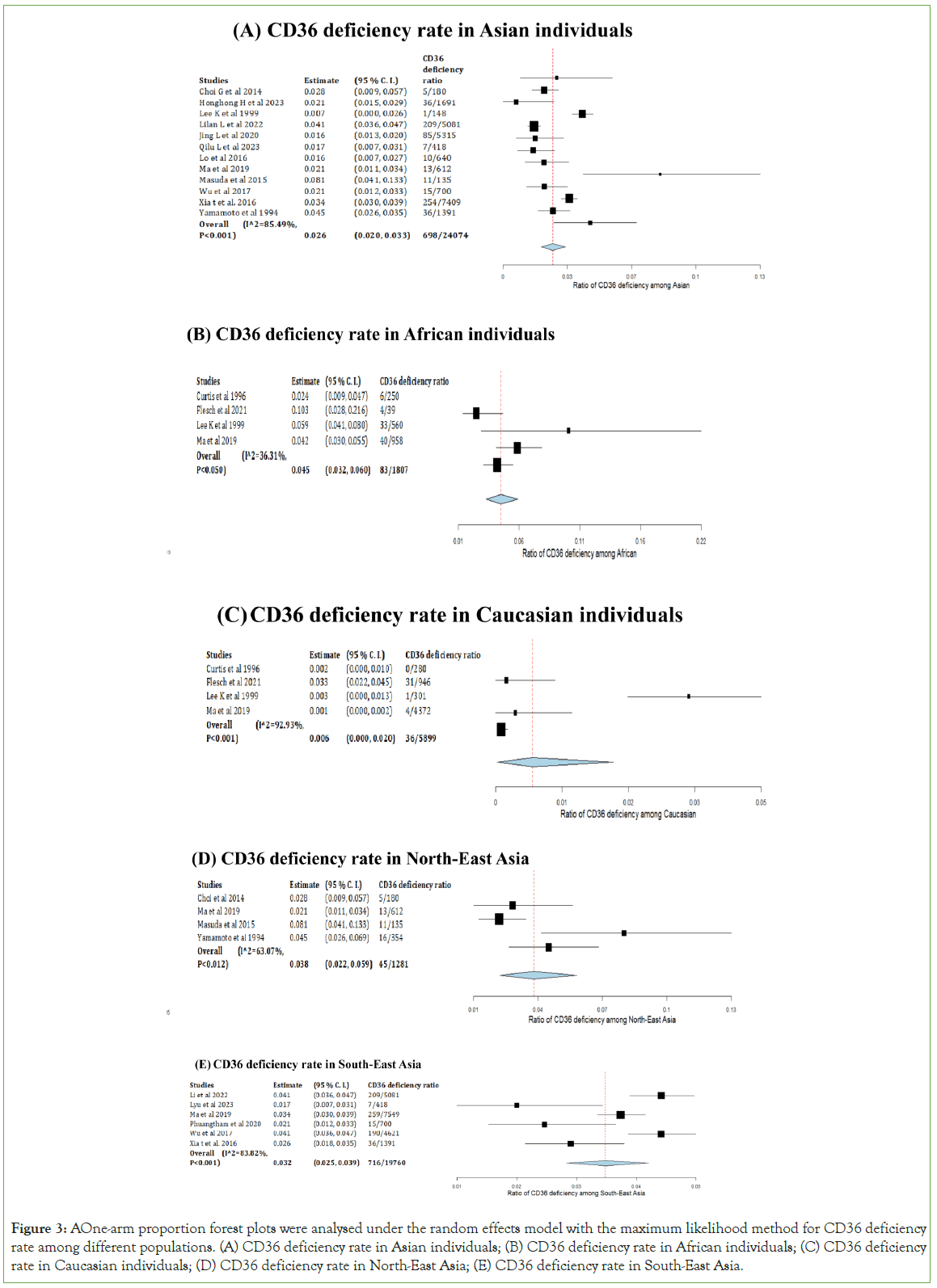
Figure 3: A One-arm proportion forest plots were analysed under the random effects model with the maximum likelihood method for CD36 deficiency rate among different populations. (A) CD36 deficiency rate in Asian individuals; (B) CD36 deficiency rate in African individuals; (C) CD36 deficiency rate in Caucasian individuals; (D) CD36 deficiency rate in North-East Asia; (E) CD36 deficiency rate in South-East Asia.
The two one-armed forest plots shown in Figures 3D and 3E, were used to analyse the incidence of CD36 deficiency among North- East and South-East Asian areas. Data from the North-East cases generated Figure 3D, with a P-value of less than 0.001, which was considered significant. The I2 was 63.07 (P-value=0.012), which indicated high discreteness in the data (95% C.I. 0.022, 0.059). The South-East individuals with CD36 deficiency were generated in Figure 3B, with a P-value less than 0.001, indicating that the overall data were significant. The high heterogeneity shown by the I2 value (83.82) and its P-value (<0.001), 95% C.I. value was 0.025-0.039. The total CD36 deficiency incidences in the North-East and South-East were 3.51% and 3.62%, respectively.
Discussion
Alloimmunisation rate caused by CD36 antibody and CD36 gene mutation rate among CD36 deficiency individuals
As mentioned, individuals with CD36 deficiency can produce antibodies that cause alloimmunisation when exposed to CD36 antibodies. Quanrui et al. reported anti-CD36 antibody-induced Fetal and Neonatal Alloimmune Thrombocytopenia (FNAIT) in Asia [24]. Another study by Xiuzhang X in China reported a case of CD36 antibody-induced Platelet Transfusion Refractoriness (PTR) and Post- Transfusion Purpura (PTP) [25]. According to previous research, not only patients who lack CD36 on platelets after transfusing normal platelets can develop platelet transfusion refractoriness, but also pregnant women who lack CD36 may develop neonatal alloimmune thrombocytopenia in newborns. Hence, one of the purposes of this systematic review and meta-analysis was to investigate the alloimmunisation and gene mutation rate caused by anti-CD36. The results provided in Figure 2A, were used in a meta-analysis to explain the alloimmunisation rate due to anti-CD36. The meta-analysis of the overall alloimmunisation ratio caused by CD36 antibodies from CD36 deficiency individuals was significant for anti-CD36 (95% C.I. 0.002, 0.027, P<0.001).
The percentage of alloimmunisation caused by anti-CD36 was 0.83% (22/2641). The rate was low; this might result from not all CD36 deficiency individuals being led alloimmunisation. The heterogeneity value was 79.71 with a P-value of less than 0.001; the high heterogeneity determined that the diversity of selected data was not caused by chance. The alloimmunisation due to anti-CD36 was 3.14% from Wu G et al. which was significantly different from the other selected data [26]. This study analysed all the alloimmunisation cases from 2008 to 2015. However, other studies included cases from healthy subjects, which might have affected heterogeneity. However, this review still had this article as its high quality. Even if the overall alloimmunisation rate is low, CD36 antibodies must still be considered during platelet transfusion or pregnancy to avoid serious consequences.
The primary reason for platelet glycoprotein deficiency is CD36 gene mutation [27]. Most alloimmunisation cases are associated with type I CD36 deficiency. The most common CD36 gene mutation type in type I CD36 deficiency occurs in three forms: C268T, 949insA, and 329-330delAC, of which C268T is the most frequent [28]. The C268T mutation indicates that there is a substitution from cysteine to threonine. This mutation affects the maturation of the CD36 88- kDa form and degrades it in the cytoplasm, further leading to silent expression of CD36 on platelets [28]. The results of the meta-analysis investigated CD36 gene mutations among individuals with platelet glycoprotein deficiency. The overall data were statistically significant with a P-value less than 0.001 (95% C.I. 0.390, 0.723), while the total percentage of CD36 gene mutations was 65.22% (120/184) in the analysed CD36 deficiency population. The heterogeneity value was 79.60 with a P-value of less than 0.001, which determined the high variability in the data. The gene mutation in the study involved thirty- five Arabian individuals, and it was 97.14% [29]. Thirty-five individuals also included a family study of one alloimmunisation case. Family members may have similar genes that can generate a high CD36 gene mutation rate, leading to a high heterogeneity value. Most of the selected studies measured CD36 deficiency individuals from the Asian and African populations. This was because of the low incidence of CD36 deficiency among Caucasian individuals. In contrast to the gene mutation ratio, the Arabian countries had the highest gene mutation (97.14%). In comparison, China (48.15%, 50.82%, and 61.11%) and Thailand (60%) had a relatively low incidence of mutations, which was lower than the overall population [9,29-32]. Generally, not all CD36 deficiencies are caused by genetic mutations. Nevertheless, CD36 deficiency in the Arabic population commonly results from genetic mutations.
CD36 deficiency among different ethnicities
Considering the time and cost for patients, it is impossible to test for CD36 deficiency and gene mutations in all the potential patients. Hence, it is essential to determine which population has a high risk of CD36 deficiency. The meta-analysis results showed the analysis of CD36 deficiency individuals of different ethnicities. Among the Asian population, the overall CD36 deficiency rate was 2.90% (698/24074) and significant in meta-analysis (95% C.I. 0.020, 0.033, P-value<0.001) with high difference among data by high heterogeneity value (I2=85.49, P-value<0.001). Furthermore, there were some differences in the African population. The overall data were significant (95% C.I. 0.032, 0.060, P-value<0.001), and the CD36 deficiency rate was relatively high (4.59%, 83/1807), I2 equal to 36.31 with P-value equal to 0.05, which indicated low heterogeneity and data were evenly distributed. Another meta-analysis was conducted in Caucasian populations. The overall results of Caucasians (0.61%, 36/5899) were significantly determined by P-value (95% C.I. 0.000, 0.020, P-value=0.013), and high heterogeneity was shown in the difference among data (I2=92.23, P-value<0.001). By comparing the platelet glycoprotein deficiency population in three major ethnicities, African individuals had the highest incidence of CD36 deficiency, and according to the study of Xu et al. the CD36 deficiency rate in Caucasians should be less than 0.4%, similar to the data in this review [27]. Overall, the Caucasian population had a low rate of CD36 deficiency, whereas African individuals had the highest rate. Therefore, patients with African backgrounds require extra awareness of platelet transfusion because they are more likely to have CD36 deficiency.
The Asian region is the largest continent, accounting for approximately 60% of the world's population [33]. The Asian data from the selected studies were separated into two categories: North-East and South-East Asia. The results of North-East Asia were statistically significant (95% C.I. 0.022, 0.059, P-value<0.001) with high heterogeneity (I2=63.07), with a P-value equal to 0.012 than the indicated variability among data. The meta-analysis results conducted in South-East Asia were significant (95% C.I. 0.025, 0.039, P-value<0.001) with I2 equal to 83.82 and P-value less than 0.001. An uneven distribution among the data was shown by a high heterogeneity value [34,35]. The overall rate of CD36 deficiency among North-East Asian (Korea, Northern China, Japan) subjects was 3.51%, while it was 3.62% for South-East Asian (Southern China, South-Eastern China, Thailand) subjects. The CD36 deficiency rate was similar between these two regions, while the South-East had a slightly higher rate [36,37]. Therefore, South-East Asians have a higher possibility of developing platelet glycoprotein deficiencies in the Asian population.
Limitations of review
Bias may have been produced from the selected studies because the study design of most selected articles was prospective, and the review was not comprehensive enough without including enough respective studies [38]. A few chosen literatures did not address the bias and limitations listed in the STROBE checklist [39-40]. The reliability of the forest plot results depended on the number of studies included, and a higher number of studies produced better representativeness. Some forest plots fit only three or four studies, which could cause high data heterogeneity, further affecting the analysis. However, the few studies in the forest plots were low due to the limited number of current studies on CD36 antibodies.
Moreover, CD36 deficiency cases included all the CD36 deficiency types instead of type II. This might have also influenced the analysed results. Finally, the meta-analysis of the CD36 gene mutation rate included only includes a few Caucasian cases, which could generate bias.
Clinical implication and future study
Alloimmunisation, especially platelet transfusion refractoriness, threatens patient well-being, necessitating careful avoidance. This review guides clinicians, emphasising the heightened susceptibility of individuals of African descent to alloimmunisation due to anti- CD36 antibodies from CD36 gene deficiency. Considering the CD36 phenotype of platelets is crucial for this population, pre-transfusion or during pregnancy. Labelling platelets with CD36 phenotype during volunteer collection is advised to prevent transfusing CD36-positive platelets to CD36-negative recipients. Furthermore, systematically exploring strategies for managing alloimmunisation induced by anti-CD36 antibodies is a prospective avenue for future inquiry. Recommending future studies with retrospective analyses enhances overall comprehensiveness in subsequent investigations.
Conclusion
This systematic review generated different meta-analyses using forest plots to investigate the alloimmunisation rate caused by anti-CD36 antibodies and which population background was most likely to cause CD36 deficiency. Overall, the research question can be answered as CD36 antibodies can cause alloimmunisation with low incidence; the Caucasian population is less able to get CD36 deficiency, while African individuals have the highest CD36 deficiency rate. The North-East and South-East Asian populations showed little difference in the CD36 deficiency distribution. The outcome can indicate to clinicians which population has a higher potential risk of producing alloimmunisation by CD36 deficiency. Investigating the varying rates of CD36 gene mutations among different ethnicities is a recommended focus for future research. Subsequent analyses should specifically target CD36 type I deficiency. Additionally, further reviews can be undertaken as more research on this subject becomes available.
Conflict of Interest
No conflicts of interest have been declared.
Funding
The authors received no financial support for the research.
References
- Hendrickson JE, Tormey CA. Understanding red blood cell alloimmunization triggers. Hematology Am Soc Hematol Educ Program. 2016;2016(1):446-451.
[Crossref] [Google Scholar] [PubMed]
- Cohn CS. Platelet transfusion refractoriness: How do i diagnose and manage? Hematology Am Soc Hematol Educ Program. 2020;2020(1):527-532.
[Crossref] [Google Scholar] [PubMed]
- Blajchman MA. Platelet transfusions: An historical perspective. Hematology Am Soc Hematol Educ Program. 2008(1):197.
[Crossref] [Google Scholar] [PubMed]
- Hod E, Schwartz J. Platelet transfusion refractoriness. Br J Haematol. 2008;142(3):348-360.
[Crossref] [Google Scholar] [PubMed]
- Stroncek DF, Rebulla P. Platelet transfusions. Lancet. 2007;370(9585):427.
[Crossref] [Google Scholar] [PubMed]
- Stanworth SJ, Navarrete C, Estcourt L, Marsh J. Platelet refractoriness–practical approaches and ongoing dilemmas in patient management. Br J Haematol. 2015;171(3):297-305.
[Crossref] [Google Scholar] [PubMed]
- Hirano K-i, Kuwasako T, Nakagawa-Toyama Y, Janabi M, Yamashita S, Matsuzawa Y. Pathophysiology of human genetic CD36 deficiency. Trends Cardiovasc Med. 2003;13(4):136-141.
[Crossref] [Google Scholar] [PubMed]
- Silverstein RL. Type 2 scavenger receptor CD36 in platelet activation: The role of hyperlipemia and oxidative stress. Clin Lipidol. 2009;4(6):767.
[Crossref] [Google Scholar] [PubMed]
- He HH, Tang LH, Jin YM, Wang YJ, Wang HM, Ding SH, et al. High-throughput CD36 phenotyping on human platelets based on sandwich ELISA and mutant gene analysis. Transfus Med Hemother. 2023;1-9
[Crossref]
- Tandon NN, Lipsky RH, Burgess WH, Jamieson GA. Isolation and characterization of platelet glycoprotein IV (CD36). J Biol Chem. 1989;264(13):7570-5.
[Crossref] [Google Scholar] [PubMed]
- Kashiwagi H, Tomiyama Y, Kosugi S, Shiraga M, Lipsky RH, Kanayama Y, et al. Identification of molecular defects in a subject with type I CD36 deficiency. Blood. 1994;83(12):3545-3552.
[Crossref] [Google Scholar] [PubMed]
- Miyaoka K, Kuwasako T, Hirano K-i, Nozaki S, Yamashita S, Matsuzawa Y. CD36 deficiency associated with insulin resistance. Lancet. 2001;357(9257):686-687.
[Crossref] [Google Scholar] [PubMed]
- Aitman TJ, Cooper LD, Norsworthy PJ, Wahid FN, Gray JK, Curtis BR, et al. Malaria susceptibility and CD36 mutation. Nature. 2000;405(6790):1015-1016.
[Crossref] [Google Scholar] [PubMed]
- Lee K, Godeau B, Fromont P, Plonquet A, Debili N, Bachir D, et al. CD36 deficiency is frequent and can cause platelet immunization in Africans. Transfusion. 1999;39(8):873-879.
[Crossref] [Google Scholar] [PubMed]
- Ma H, Shieh K-J. ELISA technique. Nat Sci. 2006;4(2):36-37.
- Brown M, Wittwer C. Flow Cytometry: Principles and clinical applications in haematology. Clin chem. 2000;46(8):1221-1229.
[Crossref] [Google Scholar] [PubMed]
- Miao G, Zhang L, Zhang J, Ge S, Xia N, Qian S, et al. Free convective PCR: From principle study to commercial applications-A critical review. Anal Chim Acta. 2020;1108:177-197.
[Crossref] [Google Scholar] [PubMed]
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. PLoS Med. 2021;18(3):e1003583.
[Crossref] [Google Scholar] [PubMed]
- Vandenbroucke JP, von Elm E, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and elaboration. Epidemiology. 2007;18(6):805-835.
[Crossref] [Google Scholar] [PubMed]
- Wallace BC, Schmid CH, Lau J, Trikalinos TA. Meta-analyst: Software for meta-analysis of binary, continuous and diagnostic data. BMC Med Res Methodol. 2009;9(1):80.
[Crossref] [Google Scholar] [PubMed]
- Choi G, Kim H, Kim S. Frequency of CD36 and CD109 deficiency on platelets: Single institute study in Korea. CYTOKINE. 2014;70(1):34.
- Curtis BR, Aster RH. Incidence of the Nak(a)-negative platelet phenotype in African Americans is similar to that of Asians. Transfusion. 1996;36(4):331-334.
[Crossref] [Google Scholar] [PubMed]
- Li L, Zhong Z, Zhou Y, Li H, Lu F, Jiang L, et al. Platelet donor database: A study on the specialist donor database for the patients with alloimmune thrombocytopenia in the Chinese population and the assessment of completeness. Chin Med J (Engl). 2022;135(2):234-236.
[Crossref] [Google Scholar] [PubMed]
- Liu QR, Liu JY, Zhao JY, Lin SB, Deng J, Xu XZ, et al. Do anti-CD36 antibodies cause only foetal/neonatal alloimmune thrombocytopenia? Br J Haematol. 2023;202(6):e62-e64.
[Crossref] [Google Scholar] [PubMed]
- Xu X, Ye X, Xia W, Liu J, Ding H, Deng J, et al. Studies on CD36 deficiency in South China: Two cases demonstrating the clinical impact of anti-CD36 antibodies. Thromb Haemost. 2013;110(6):1199-1206.
[Crossref] [Google Scholar] [PubMed]
- Wu G, Zhou Y, Li L, Zhong Z, Li H, Li H, et al. Platelet immunology in China: Research and clinical applications. Transfus Med Rev. 2017;31(2):118-125.
[Crossref] [Google Scholar] [PubMed]
- Xu X, Zheng X, Zhu F. CD36 gene variants and their clinical relevance: A narrative review. Ann Blood. 2021;6:34.
- Rac ME, Safranow K, Poncyljusz W. Molecular basis of human CD36 gene mutations. Mol Med. 2007;13(5):288-296.
[Crossref] [Google Scholar] [PubMed]
- Flesch BK, Scherer V, Opitz A, Ochmann O, Janson A, Steitz M, et al. Platelet CD36 deficiency is present in 2.6% of Arabian individuals and can cause NAIT and platelet refractoriness. Transfusion. 2021;61(6):1932-1942.
[Crossref] [Google Scholar] [PubMed]
- Xia W, Xu X, Fu Y, Ye X, Tsuno N, Santoso S, et al. CD36 deficiency among South-East Asian populations. ISBT Sci Series. 2016;11(S2):33-36.
- Phuangtham R, Santoso S, Leelayuwat C, Komvilaisak P, Ding H, Romphruk AV. Frequency of CD36 deficiency in Thais analyzed by quantification of CD36 on cell surfaces and in plasma. Transfusion. 2020;60(4):847-854.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Shao Y, Ding H, Deng J, Xu X, Wang J, et al. Distribution of CD36 deficiency in different Chinese ethnic groups. Hum Immunol. 2020;81(7):366-371.
[Crossref] [Google Scholar] [PubMed]
- Goerres A, Vanhuysse P. Global political demography the politics of population change. Cham Spring I Publish AG. 2021.
- Ohto H, Miura S, Ariga H, Ishii T, Fujimori K, Morita S. The natural history of maternal immunization against foetal platelet alloantigens. Transfus Med. 2004;14(6):399-408.
[Crossref] [Google Scholar] [PubMed]
- Lee K, Godeau B, Fromont P, Plonquet A, Debili N, Bachir D, et al. CD36 deficiency is frequent and can cause platelet immunization in Africans. Transfusion. 1999;39(8):873-879.
[Crossref] [Google Scholar] [PubMed]
- Lyu Q, Peng M, Chen Q, Ji X, Wang Z, Li Q, et al. Frequency and molecular basis of CD36 deficiency among platelet donors in Kunming, China. Platelets. 2023;34(1):2176168.
[Crossref] [Google Scholar] [PubMed]
- Lo SC, Lin KH, Hsieh HH, Lin DT, Hu CY. Genetic variations of CD36 and low platelet CD36 expression - a risk factor for lipemic plasma donation in Taiwanese apheresis donors. Vox Sang. 2016;110(3):236-243.
[Crossref] [Google Scholar] [PubMed]
- Ma C, Wang J, Yang L, Feng Y, Fu L, Guan X, et al. A single-centre investigational study of CD36 antigen deficiency and platelet alloantibody distribution in different populations in Northern China as well as platelet alloantibodies effect on pregnancy. Clin Chim Acta. 2019;498:68-75.
[Crossref] [Google Scholar] [PubMed]
- Masuda Y, Tamura S, Matsuno K, Nagasawa A, Hayasaka K, Shimizu C, et al. Diverse CD36 expression among Japanese population: Defective CD36 mutations cause platelet and monocyte CD36 reductions in not only deficient but also normal phenotype subjects. Thromb Res. 2015;135(5):951-957.
[Crossref] [Google Scholar] [PubMed]
- Yamamoto N, Akamatsu N, Sakuraba H, Yamazaki H, Tanoue K. Platelet glycoprotein IV (CD36) deficiency is associated with the absence (type I) or the presence (type II) of glycoprotein IV on monocytes. Blood. 1994;83(2):392-397.
[Crossref] [Google Scholar] [PubMed]
Citation: Wang Y, Jackson DE (2024) Incidence of CD36 Antibody Induced Alloimmunisation and CD36 Deficiency Distribution among Different Populations: A Systematic Review and Meta-Analysis. J Blood Disord Transfus. 15.578.
Copyright: © 2024 Wang Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.