Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 10, Issue 8
Identification of Stripe Rust Resistance in Ethiopian Durum Wheat by Phenotypic Screening and Kompetitive Allele Specific PCR (KASP) SNP Markers
Sisay Kidane Alemu1*, Ayele Badebo2, Kassahun Tesfaye3 and Cristobal Uauy42CIMMYT, Addis Ababa, Ethiopia
3Institute of Biotechnology, Addis Ababa University and DMCMB, Addis Ababa, Ethiopia
4John Innes Centre, Norwich, UK
Received: 18-Aug-2019 Published: 29-Nov-2019, DOI: 10.35248/2157-7471.19.10.483
Abstract
Stripe (Yellow) rust caused by Puccinia striiformis f.sp. tritici (Pst) is one of the most devastating diseases of wheat in the highlands of Ethiopia. Improved cultivars often lose their resistance due to occurrence of new virulent races which overcome the genes and make the cultivars out of production. Therefore, identification of new sources of resistance genes helps in battling yellow rust and maximizes wheat production in Ethiopia. In this study, 300 durum wheat lines (landraces & cultivars) were screened with three virulent isolates (Pst_Is1, Pst_Is4 and Pst_Is8) for seedling resistance using Infection Type (IT) scoring method. The lines were also screened with 16 KASP-based SNP markers linked to 7 Yr genes already identified in various studies. Highly resistant infection type (IT: 0 -3) to Pst_Is1, Pst_Is4, and Pst_Is8 was exhibited by 59.3%; 67.3%; and 46.3% of the lines, respectively. 124 lines constantly exhibited high level of resistance to all three isolates. The majority (96.8%) of the resistant lines are landraces while four (3.2%) are commercial cultivars (Cocorit/71, Yerer, Obsa and Dire). In the molecular screening 12 of the markers gave clear amplifications in the controls and the tested lines. Yr7, Yr15 and YrSp were detected in 81.7%, 88.3% and 0.7% of the lines respectively while Yr1, Yr17 and Yr36 were not detected. Detection frequency was higher in landraces (58.7%) than in cultivars (32.8%). Gene combinations frequency was the highest (72.7%) for Yr7+Yr15 followed by Yr15+YrSp (0.3%). Overall, this study has resulted in detection of genes Yr15 and YrSp, which are potential candidates for marker assisted breeding for Pst resistance in wheat. Besides, it has shown that resistant source identification and detection of genes can be facilitated through combined application of phenotyping and molecular screening.
Keywords
Resistance gene; Screening; Molecular markers; KASP; SNP; Yellow rust; Infection type
Introduction
Wheat is among the most important staple food crops in Ethiopia produced at 1.69 million ha of land with an annual yield approximated to 4.64 million metric ton [1]. Compared to maize, teff and sorghum, it is the third and fourth important crop in terms of production and production area respectively [1].
Stripe (Yellow) rust caused by Puccinia striiformis f.sp. tritici (Pst) is one of the most important diseases of wheat causing serious damage to wheat production in the highlands of Ethiopia [2-4]. The disease causes substantial yield and grain quality reduction under severe infestations [2,5]. Varietal susceptibility, severity of infection, low temperature and altitude are among the main factors determining the extent of losses. Severe losses up to 60% can be observed in terms of poor grain quality and damaged tillers [6]. Under very favorable conditions for the pathogen, especially at high altitude, damages up to 100% crop loss are common [6]. In Ethiopia, frequent yellow rust epidemics have occurred in the past accompanied by appearance of virulent races causing susceptibility on popular bread wheat verities like Lakech [7] and Dashen [8]. A devastating yellow rust epidemic occurred in the year 2010, was projected to have infected over 400,000 ha of wheat which led to an estimated expense of more than US$3.2 million on fungicides [9].
Disease management methods such as, cultural practices, fungicide application and resistance genes/cultivars deployment are integral part of wheat production with the latter being the most preferred management strategy because it is economical and environmentally safe [10-13]. However, due to the neverending host-pathogen interactions, resistance genes often succumb to emerging Pst races soon after commercialization and that calls for continuous search and identification of new sources of resistance to sustain wheat production. The fact that Ethiopia is among the secondary centre of diversity [14-16] to tetraploid wheat is a natural endowment to look for various traits of interest including rust resistance. Sources of resistance to Pst are identified through both phenotypic and molecular screening of host genotypes.
The phenotypic screening approach requires knowledge on the virulence status of prevailing races in the pathogen population and the reactions of the corresponding host genotypes. Virulence surveys are of prime importance to trace Pst population dynamics and identify emerging races [17-19]. Besides, rust samples are sent to the Global Rust Reference Center (GRRC) somehow on regular basis so that their virulence status is determined (http://wheatrust.org/). Such Pst races of known virulence are used in screening germplasm both at adult and seedling stage to identify resistant lines [8,20]. The identified resistance sources are either directly advanced to variety trials or used as parental lines in crossing programs for development of resistant cultivars.
Molecular marker-based screening of resistance sources with diagnostic or linked markers is an alternative to the phenotypic approach for detection of the presence or absence of resistant genes in the genotypes [21,22]. A wide range of molecular markers (SSR, RFLP, STS, SCAR, CAPS, RAPD, DArt) are reported to be linked or diagnostic to most of the Yr genes in wheat [23,34]. However, there are limited progress in the development of diagnostic/linked KASP based SNP markers for the Yr genes and they are few in number compared to the SSRs and other markers systems [25-40].
Detection of Yr genes with linked molecular markers in Ethiopian bread wheat cultivars was reported by Dawit et al. [41] although similar works on durum wheat are scarce. KASP based assays are relatively easy-going, have better resolution power [42] and low cost effective [43] as compared to SSRs and other PCR based markers. In Ethiopia, several germplasm evaluation activities for resistance to stripe rust are carried out in the field and greenhouse using phenotypic screening.
Molecular screening approach together with the phenotyping method provides complementary evidence for the reliable identification of resistance genes. Therefore, this study was conducted with the following objectives: to identify resistance in Ethiopian durum wheat (landraces and cultivars) through phenotypic screening using characterized Pst isolates and to detect known Yr genes through molecular screening using KASPbased SNP markers linked to already reported Yr genes.
Materials and Methods
The wheat materials and Stripe rust pathogen
Three hundred durum wheat lines were used for the study. They were composed of 261 landraces which are maintained at Ethiopian Biodiversity Institute (EBI), Debre-Zeit Research Center (DzARC) and Ethio-Organic Seed Action (EOSA) while 39 cultivars were obtained from DzARC. All the lines were grown for two rounds of self-pollination through single seed descent method to have a relatively pure starting material. Yr single-gene differentials and Avocet-S variety were included as positive and negative controls for the respective Yr genes in the molecular screening. Three virulent stripe rust isolates (Pst_Is1, Pst_Is4 and Pst_Is8) selected from Pst samples (collected from Meraro, Kulumsa, and Chefe-Donsa) were used for the phenotypic screening.
Screening of durum lines for their resistance at seedling stage
A total of 300 durum wheat lines (landraces and cultivars) were inoculated with three virulent isolates of yellow rust (Table 1) at seedling stage in the greenhouse facility at Kulumsa research center. The isolates were selected from ten Pst samples originally collected from three testing sites (Meraro, Kulumsa and Chefe- Donsa). The selection was based on virulence test carried out on a set of 35 stripe rust differential lines corresponding to 19 resistance genes: Yr1, Yr2, Yr3, Yr4, Yr5, Yr6, Yr7, Yr8, Yr9, Yr10, Yr15, Yr17, Yr24, Yr25, Yr27, Yr32, YrSp, YrAvS and Yr Amb. List of the tested lines with some descriptive information is presented. The three virulent isolates (Pst_Is1, Pst_Is4 and Pst_Is8) were maintained on cultivar Morocco. Five seeds of each line were sown in 7 × 7 cm black square plastic pots filled with soil: compost: sand in a 2:1:1 (v/v/v) ratio and allowed to grow in a greenhouse compartment at 16-18°C. Susceptible durum (local-red and LD357) and a bread wheat (Morocco) cultivars were included in the test to serve as positive controls for successful inoculation and infection establishment. A weeklater, nine pots of seedlings were placed together on a sample tray and inoculated with ~3.5 mg of fresh spores suspended in 0.25 ml of mineral oil in gelatin capsule using vacuum pump sprayer. After drying the oil for about five minutes in an open air, the seedlings were moisturized with a fine spray of water, incubated at temperature 8-10°C and 100% RH for 24 hours of darkness. The seedlings were then transferred to greenhouse compartment with temperature 18-22°C. Seedling reaction in terms of Infection Type (IT) was evaluated 14-16 days after inoculation using 0-9 scale of McNeal et al., [44]. Lines with 0-3 IT score were considered as resistant, 4-6 as intermediate and 7-9 as susceptible [44]. The test was performed in four replications for Pst_Is1 and Pst_Is4 while in two replications for Pst_Is8 over time taking one isolate at a time to avoid cross contamination. Frequencies of various response groups were determined by analyzing the number of lines having resistant (IT:0-3), Intermediate (IT:4-6) and susceptible (IT:7-9) reactions among the tested lines. The durum wheat lines with resistant reactions to all three isolates were also identified by aligning the respective IT values of each line across isolates. Besides, response groups within and between landraces and cultivars were also compared to identify the relative richness of the lines for Pst resistance.
| PST. Sample name | Sample Code | Virulence/Avirulence formula for Pst isolates tested on 19 single Yr gene differentials |
|---|---|---|
| Pst_Is1 | ET_Or_KARC_L.Red_2015_1 | 1,2,4,6,7,8,9,17,27,32, YrAvS, YrAmb/3+,5,10+, 15, 24, 25, SP |
| Pst_Is4 | ET_Or_Mer_EDW_262_2015 | 2,3+,4,6,7,8,9,10+,17, 24, 25, 27, 32, YrAvS/1,5, 15, SP, YrAmb |
| Pst_Is8 | ET_Or_Mer_EDW_270_2015 | 1,2,3+,6,7,8,9,10+,17, 24, 25, 27, 32, YrAvS, YrAmb/4, 5, 15, SP |
| *Yr-gene differentials used for differentiating the pst samples were obtained from GRRC, Denmark and listed as follows: 1=Chinese 166 (Yr1), 2=Kalyansona (Yr2), 3=Vilmorin 23 (Yr3+), 4=Hybrid 46 (Yr4), 5=Avocet Yr5 (Yr5), 6=Avocet Yr6 (Yr6), 7=Avocet Yr7 (Yr7), 8=Avocet Yr8 (Yr8), 9=Avocet Yr9 (Yr9), 10=Moro (Yr10+), 15=Cortez (Yr15), 17=Avocet Yr17 (Yr17), 24=Avocet Yr24 (Yr24), 25=Avocet Yr25 (Yr25), 27=Avocet Yr27 (Yr27), 32=Avocet Yr32 (Yr32), SP=Avocet YrSp (YrSp), Avocet S=YrAvS and Ambition=YrAmb. | ||
Table 1: Stripe rust (Puccinia striiformis f.sp. tritici ) isolates used for phenotyping assessment.
Molecular screening for known Yr genes
DNA extraction and quantification: Wheat seeds were germinated in Petri dishes with 1 mm layer of water. They were incubated at 4°C for 12 hrs and further transferred to room temperature (25°C) for 3-4 days. Uniformly germinated seedlings were transferred to a 96 cupped black plastic trays filled with Peat (85%) & Sand (15%) Mix and incubated in cereal growth chamber. The incubation conditions were 100% (1000 or 1200 μmol m-2 s-1) light intensity; RH of 70%; photoperiod of 16 h light at 19°C and 8 h dark at 16°C temperatures. Genomic DNA was extracted from 10-14 days old seedlings following the wheat DNA extraction protocol in 96-well plates (http://www.wheattraining. com) [45]. DNA quantification was carried out using NanodropTM (8-sample spectrophotometer ND-800, ThermoFischer Scientific, USA) and normalized to ~20 ng/μL for each sample.
Genotyping with Kompetitive allele specific PCR (KASP) assay: Molecular screening of the tested lines was conducted through haplotype analysis of 16 KASP-based SNP markers linked to selected seven Yr genes. Selection of the marker was based on literature from similar studies on virulence survey of Pst races, recommendations and availability of KASP assay SNP markers linked to the respective genes (Table 2). The primers sequences, along with the attached FAM (5 ’ GAAGGTGACCAAGTTCATGCT 3 ’ ) or HEX (5 ’ GAAGGTCGGAGTCAACGGATT 3’) tails were ordered from Sigma-Aldrich. A final volume of 100 μL primer mix was prepared following the manufacturer ’ s (LGC Group, Teddington, United Kingdom) instructions as 12 μL of each of the tailed forward primer (100 μM), 30 μL common primer (100 μM) and toped up by 46 μL distilled water [46].
| Gene | Chr | Assay ID/ Primer name | Primer_Sequences (5' to 3') | FAM-allele | HEX-allele | Reference |
|---|---|---|---|---|---|---|
| Yr1 | 2AL | IWB44454/ A1 primer | GAAGGTGACCAAGTTCATGCTGGCTGACATGTGTTCCAGACACT | T | Bansal et al. [38] | |
| IWB44454/ A2 primer | GAAGGTCGGAGTCAACGGATTGCTGACATGTGTTCCAGACACG | G | ||||
| IWB44454/ Common | CTGGGGCCTCGGGAGATTTTGAA | |||||
| Yr1 | 2AL | IWB81533/A1 primer | GAAGGTGACCAAGTTCATGCTGTTTGCTGTCGTCGAGGAGCTT | T | Bansal et al. [38] | |
| IWB81533/A2 primer | GAAGGTCGGAGTCAACGGATTTGCTGTCGTCGAGGAGCTC | C | ||||
| IWB81533/ Common | CCCAGTGGGAGATCTCCACCTT | |||||
| Yr1 | 2AL | IWB44619/A1 primer | GAAGGTGACCAAGTTCATGCTGGAGTATTACTTGCTGGACCCTA | A | Bansal et al. [38] | |
| IWB44619/A2 primer | GAAGGTCGGAGTCAACGGATTGGAGTATTACTTGCTGGACCCTC | C | ||||
| IWB44619/ Common | AAGGCTCTGAACAATGAACTTGCTGTAT | |||||
| Yr5 | 2B | O_LYr5CadCl | GAAGGTCGGAGTCAACGGATTGATACTGGATGACAAAATTTTTA | A | Marchal et al. [40] | |
| O_Other | GAAGGTGACCAAGTTCATGCTGATACTGGATGACAAAATTTTGC | C | ||||
| O_Common | GGTTTTTCAGATTATGGAACA | |||||
| Yr5 | 2B | T_Common | GAAGGTGACCAAGTTCATGCTGTTGTTGCACTTTACAAATCCA | A | Marchal et al. [40] | |
| T_LYr5_others | GAAGGTCGGAGTCAACGGATTGTTGTTGCACTTTACAAATCCC | C | ||||
| T_CadCl | CGGTTTCTGGATGTCACA | |||||
| YrSP | 2B | KASP_Yr5 | GAAGGTCGGAGTCAACGGATTGAGAAAATCAGCAGGTGC | C | Marchal et al. [40] | |
| KASP_YrSP | GAAGGTGACCAAGTTCATGCTGAGAAAATCAGCAGGTGG | G | ||||
| KASP_Common | AGCGAGTTGAGGACATTGGT | |||||
| Yr7 | 2B | AL_C7_R | GAAGGTCGGAGTCAACGGATTTTAGTCCTGCCCCATAAGCG | G | Marchal et al. [40] | |
| AL_Alt_R | GAAGGTGACCAAGTTCATGCTTTAGTCCAGCCCCATAAGCC | C | ||||
| AL_Com_F | CAGTGTTAAAACCAGGGAGGA | |||||
| Yr7 | 2B | AR_C7_F | GAAGGTCGGAGTCAACGGATTTGGAGGTATCATCTGGTGAG | G | Marchal et al. [40] | |
| AR_Alt_F | GAAGGTGACCAAGTTCATGCTTGGAGGTATCATCGGGTGAA | A | ||||
| AR_Com_R | CATCAAAATCATCGCCTATGT | |||||
| Yr7 | 2B | AX_C7_R | GAAGGTCGGAGTCAACGGATTCACATGAGTCGATACTGAGG | G | Marchal et al. [40] | |
| AX_Alt_R | GAAGGTGACCAAGTTCATGCTCACACGACCTAATACTGAGA | A | ||||
| AX_Com_F | ACTGCAATGCCTTCCCATA | |||||
| Yr7 | 2B | AAN_C7_F | GAAGGTCGGAGTCAACGGATTGCTGGAAAGGCTTGACATCA | A | Marchal et al. [40] | |
| AAN_Alt_F | GAAGGTGACCAAGTTCATGCTGCTGGAAAGGCTTGAGATCG | G | ||||
| AAN_Com_R | AATGGCGTGGTAAGGACAGA | |||||
| Yr15 | 1BS | Yr15-R5 | GAAGGTGACCAAGTTCATGCTAGTCAACTTGGATTACACTGAAGTT | T | Ramirez-Gonzalez et al. [39] | |
| GAAGGTCGGAGTCAACGGATTAGTCAACTTGGATTACACTGAAGTC | C | |||||
| AGATATCACACTGAACATACTGATGAG | ||||||
| Yr15 | 1BS | Yr15-R8 | GAAGGTGACCAAGTTCATGCTCAGATCCCCGGTTCTCTCAAG | G | Ramirez-Gonzalez et al. [ 39] | |
| GAAGGTCGGAGTCAACGGATTCAGATCCCCGGTTCTCTCAAA | A | |||||
| CCCCCAAATGATCGAGAATA | ||||||
| Yr15 | 1BS | Yr15-R11 | GAAGGTGACCAAGTTCATGCTCCATTCTGATCAAGGTCACTGTCG | G | Ramirez-Gonzalez et al. [ 39] | |
| GAAGGTCGGAGTCAACGGATTCCATTCTGATCAAGGTCACTGTCA | A | |||||
| TTCTGTATGGCAACGGGAGC | ||||||
| Yr17 | 2AS | VPM_SNP_AL1 | GAAGGTGACCAAGTTCATGCTCGCCGTTCCGAAYACGAGA | A | Helguera et al. [26] | |
| VPM_SNP_AL2 | GAAGGTCGGAGTCAACGGATTCGCCGTTCCGAAYACGAGG | G | ||||
| VPM_SNP_C | CCCTGGCTTGCACCTTCGACAA | |||||
| Yr17 | 2AS | Lr37_AL1 | GAAGGTGACCAAGTTCATGCTGGACGGCGTTTGCTCATGCTA | A | Helguera et al. [26] | |
| Lr37_AL2 | GAAGGTCGGAGTCAACGGATTAGGACGGCGTTTGCTCATGCTG | G | ||||
| Lr37_C1 | AGCAGTATGTACACAAAA | |||||
| Yr36 | 6AS | wMAS000017 | GAAGGTGACCAAGTTCATGCTCAAGAGGGGAGAGACATGTTACTTA | A | Distelfeld et al. [36], | |
| GAAGGTCGGAGTCAACGGATTCAAGAGGGGAGAGACATGTTACTTT | T | Uauy et al. [35], Fu et al. [39] | ||||
| GATTATGGGAGTAGGTTGGTGAGATAAAA |
Table 2: KASP assay primers used in screening of Ethiopian durum wheat germplasm for detection of Yr genes linked to the corresponding SNPs.
The KASP assay for the corresponding SNP markers were tested using a subset of 24 lines including the positive and negative controls of the respective Yr genes. The positive controls are Yr gene containing differential lines and the negative control is a susceptible wheat line called Avocet-S without any of the Yr genes. The reaction was set up as final reaction volume of 5.07 μL which is composed of 0.07 μL of primer mix, 2.5 μL of KASP master mix to which 2.5 μL template (50 ng) DNA was added. The thermocycling conditions was carried out in Eppendorf Mastercycler pro 384 using the optimized program at Uauy’s Lab JIC as follows: hot start at 95°C for 15 min, ten touchdown cycles (95°C for 20 s; touchdown at 65°C; decreasing 1°C per cycle; 25 s) which is followed by 30 cycles of amplification (95°C for 10 s; 57°C; 60 s). At the end of the 30 cycles, plates were read on PHERASTAR plate reader (BMG LABTECH, Germany). Under condition of weak signal and no genotype cluster formation, an additional 5 cycle of PCR was performed, and reading was recorded again. Once all the KASP assays for all the markers were confirmed, amplification of whole samples was performed at the KASP genotyping service unit available at John Innes Centre (JIC) Norwich. Data analysis was performed using Klustercaller software (version 2.22.0.5; LGC Hoddesdon, UK).
Segregation of the genotyped lines was examined relative to the positive control alleles for the respective Yr genes. Lines which had allele amplification the same as the positive controls and segregated together were considered as having the Yr gene. Finally, detection frequency of the markers, proportion of tested lines having a single and combination of the detected Yr genes were investigated across the tested lines.
Results
Reaction of the landraces and cultivars to Pst isolates
Various levels of reactions observed among the 300 durum wheat lines to the three Pst Isolates. The number and proportion of durum wheat lines in different resistance classes across the three isolates is summarized in Table 3. In total, 178 (59.3%), 202 (67.3%) and 139 (46.3%) of the lines were resistant (IT: 0 - 3) while 36 (12%), 23 (7.7%) and 51 (17%) were susceptible (IT:7 - 9) to Pst_Is1, Pst_Is4 and Pst_Is8 respectively. Within the landraces, IT averaged 3.07 ± 0.11, 2.77 ± 0.09 and 3.67 ± 0.11 for Pst_Is1, Pst_Is4 and Pst_Is8 respectively. Within the cultivars however, the average IT was 5.75 ± 0.31, 5.25 ± 0.28 and 5.96 ± 0.33 for Pst_Is1, Pst_Is4 and Pst_Is8 respectively.
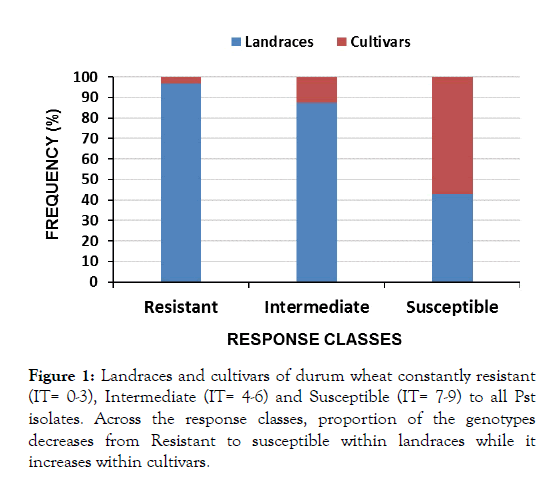
Of all the lines, a total of 170 lines appeared commonly across the three isolates of which 124 (72.9%), 32 (18.8%) and 14 (8.2%) were resistant, intermediate and susceptible respectively (Table 3 and Figure 1). Among the resistant lines, 120 (96.8%) were landraces and 4 (3.2%) were cultivars. The four cultivars resistant across all isolates are Cocorit/71, Yerer, Obsa and Dire.
Figure 1: Landraces and cultivars of durum wheat constantly resistant (IT= 0-3), Intermediate (IT= 4-6) and Susceptible (IT= 7-9) to all Pst isolates. Across the response classes, proportion of the genotypes decreases from Resistant to susceptible within landraces while it increases within cultivars.
| Number of genotypes (%) | ||||
|---|---|---|---|---|
| Isolate (Location)* | Resistance Class (IT)** | Landraces | Cultivars | Total |
| Pst_Is1 (KARC) | R (0-3) | 173 (66.3) | 5 (12.8) | 178 (59.3) |
| I (4-6) | 69 (26.4) | 17 (43.6) | 86 (28.7) | |
| S (7-9) | 19 (7.3) | 17 (43.6) | 36 (12) | |
| Total | 261 (100) | 39 (100) | 300 (100) | |
| Pst_Is4 (Meraro) | R (0-3) | 194 (74.3) | 8 (20.5) | 202 (67.3) |
| I (4-6) | 58 (22.2) | 17 (43.6) | 75 (25) | |
| S (7-9) | 9 (3.4) | 14 (35.9) | 23 (7.7) | |
| Total | 261 (100) | 39 (100) | 300 (100) | |
| Pst_Is8 (Meraro) | R (0-3) | 134 (51.3) | 5 (12.8) | 139 (46.3) |
| I (4-6) | 99 (37.9) | 11 (28.2) | 110 (36.7) | |
| S (7-9) | 28 (10.7) | 23 (59) | 51 (17) | |
| Total | 261 (100) | 39 (100) | 300 (100) | |
| Across all Isolates | R (0-3) | 120 (82.8) | 4 (25) | 124 (72.9) |
| I (4-6) | 28 (19.3) | 4 (25) | 32 (18.8) | |
| S (7-9) | 6 (4.1) | 8 (50) | 14 (8.2) | |
| Total | 154 (100) | 16 (100) | 170 (100) | |
| *The three Pst isolates used are selected from virulence test of 10 isolates (Table S1) on differentials corresponding to 19 Yr genes and they are avirulent to Yr5, Yr15 & YrSp but virulent to all the rest. ** IT=Infection Type, was evaluated from 0 - 9 scale as described by McNeal et al., (1971) [44] where R=Resistant, I=Intermediate and S=Susceptible | ||||
Table 3: Number and percentage of Landrace and cultivars of durum wheat in classes of resistance to three Pst isolates.
Molecular screening
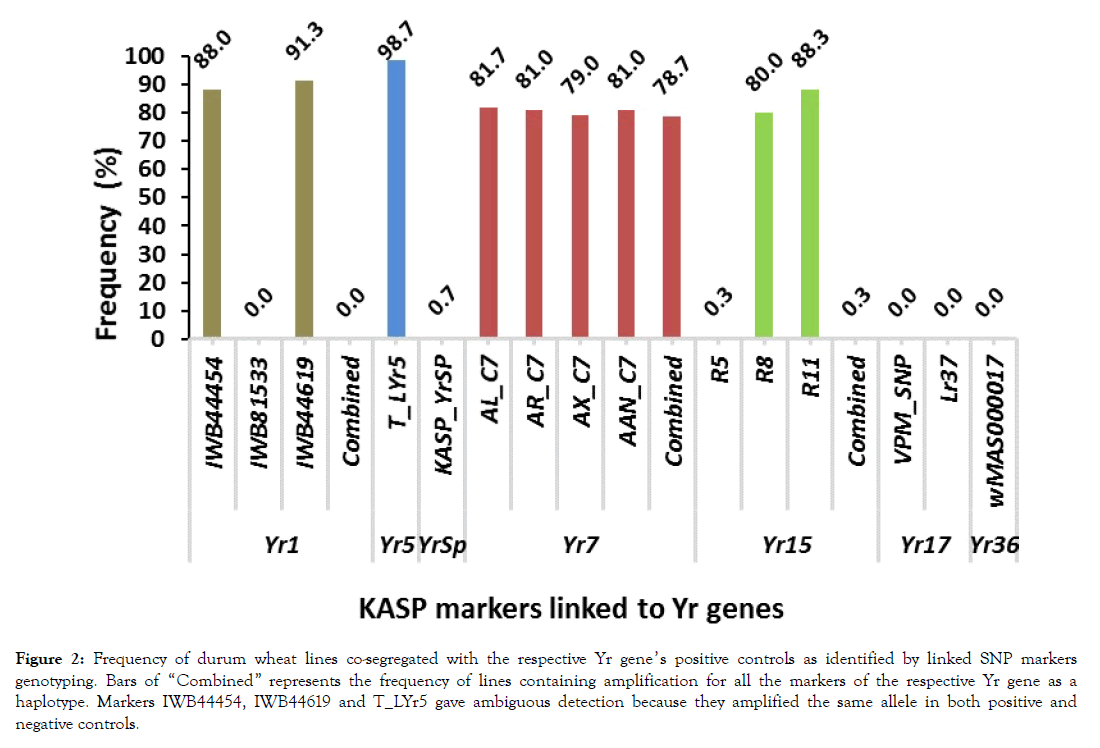
Detection of Yr genes amongst tested lines: The KASP genotyping summary for the screening of the 300 lines and positive and negative controls with 16 SNP markers linked to the 7 already reported Yr genes and frequency of lines which had amplifications with markers is presented in Table 4 and Figure 2. The presence of Yr1 was assayed with three KASP SNP markers (IWB44454, IWB81533 and IWB44619) of which, IWB81533 amplified the Fam_allele-T in the positive control (AvSYr1NIL) and the Hex_allele-C in the negative control (Avocet-S). All the 300 lines had amplification for the Hex_allele- G and segregated with the negative control indicating Yr1 is not detected. Amplifications with the other two markers (IWB44454 and IWB44619) were not contrasting between the controls. Assay for presence of Yr5 was carried out with markers O_LYr5CadCl and T_LYr5. For marker O_LYr5CadCl, no amplification was found for both controls and tested lines as well (Table 4). Marker T_LYr5 on the other hand gave amplification for only the Hex_allele-C in both controls and in 296 (98.7%) of the tested lines which was an ambiguous detection (Table 4 and Figure 2). Marker KASP_YrSp, which was used to detect YrSp, amplified the Hex_allele-C in the positive control and only in two of the tested lines (0.7%) while no amplification was found in the negative control and in almost all the tested lines (Table 4 and Figure 2). All the four markers (AL_C7, AR_C7, AX_C7 and AAN_C7) applied for detection of Yr7 perfectly amplified the Hex_ and F am_ alleles in the positive and negative controls respectively and in the tested lines as well (Table 4, Figure 2 and Figures 3B to 3E). With the best amplifying linked marker (AL_C7) the gene was detected in 81.7% of the lines although the combined amplification all the four markers gave detection in 78.7% of the lines (Figure 2). The detection of Yr15, performed by markers R5 amplified the Fam_allele-C in the positive controls and in just one of the tested lines. For R8, Fam-allele-G was amplified in the positive control and in 57 of the tested lines. Marker R11 appeared as the best amplifying one and resulted in the detection of the gene in 88.3% of the teste lines. When all three markers are considered in combined detection of Yr15, it was found in only 0.3% of the lines (Figure 2 and Figures 3G-3I). Presence of Yr17 was assessed with marker VPM_SNP and leaf rust resistant gene related marker Lr37. Both VPM_SNP and Lr35 amplified their respective positive and negative control alleles. Whereas only the negative control alleles were amplified among the tested lines and no Yr17 was detected (Table 4 and Figure 2). Similarly, existence of Yr36 was examined with marker wMAS000017. Hex_allele-T and the negative Fam-allele-A were amplified the positive and negative controls respectively while only the negative control allele was amplified among the tested lines and here again no Yr36 was detected.
| Marker name | Gene | FAM-allele | HEX-allele | Genotype call of Controls* | Number of lines per genotype calls | Detection Condition** | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | YY | YX | XX | DNA | |||||
| IWB44454 | Yr1 | T | G | XX (TT) | XX (TT) | 20 | 1 | 264 | 15 | Ambiguous |
| IWB81533 | Yr1 | T | C | XX (TT) | YY (CC) | 300 | 0 | 0 | 0 | Contrasting |
| IWB44619 | Yr1 | A | C | YY (CC) | YY (CC) | 274 | 0 | 21 | 5 | Ambiguous |
| O_LYr5CadCl | Yr5 | C | A | DNA (--) | DNA (--) | 5 | 0 | 18 | 277 | NA |
| T_LYr5 | Yr5 | A | C | YY (CC) | YY (CC) | 296 | 3 | 0 | 1 | Ambiguous |
| KASP_YrSP | YrSp | G | C | YY (CC) | DNA (--) | 2 | 0 | 2 | 296 | Contrasting |
| AL_C7 | Yr7 | C | G | YY (GG) | XX (CC) | 245 | 0 | 52 | 3 | Contrasting |
| AR_C7 | Yr7 | A | G | YY (GG) | XX (AA) | 243 | 0 | 57 | 0 | Contrasting |
| AX_C7 | Yr7 | A | G | YY (GG) | XX (AA) | 237 | 3 | 59 | 1 | Contrasting |
| AAN_C7 | Yr7 | G | A | YY (AA) | XX (GG) | 243 | 1 | 54 | 2 | Contrasting |
| R5 | Yr15 | T | C | YY (CC) | XX (TT) | 1 | 1 | 296 | 2 | Contrasting |
| R8 | Yr15 | G | A | XX (GG) | YY (AA) | 57 | 1 | 240 | 2 | Contrasting |
| R11 | Yr15 | G | A | YY (AA) | XX (GG) | 265 | 2 | 32 | 1 | Contrasting |
| VPM_SNP | Yr17 | A | G | XX (AA) | YY (GG) | 297 | 0 | 0 | 3 | Contrasting |
| Lr37 | Yr17 | A | G | YY (GG) | XX (AA) | 0 | 0 | 290 | 10 | Contrasting |
| wMAS000017 | Yr36 | A | T | YY (TT) | XX (AA) | 0 | 0 | 295 | 5 | Contrasting |
| *Positive controls used for the respective Yr genes are described as follow: Yr1= AvSYr1NIL, Yr5= AvSYr5NIL, Yr7= AvSYr7NIL, Yr15= AvSYr15NIL, Yr17= AvSYr17NIL, Yr36= UC_GPC_B1_P_3 and YrSP=AvSYrSpNIL. Avocet S for negative control. UC_GPC_B1_P_3 is spring wheat having Grain Protein Content (GPC) gene linked to Yr36 and is obtained from Uauy lab Crop Genetics department, JIC. In the genotype call, X is the Fam-allele call and Y is the HEX-allele call. ** Ambiguous diagnostic value means: genotype call of the Positive and Negative controls is the same making detection of the tested genotypes non-conclusive; and NA is for the marker which did not amplify. | ||||||||||
Table 4: KASP genotyping summary of durum wheat lines screened with 16 SNP markers linked to seven Yr genes.
Figure 2: Frequency of durum wheat lines co-segregated with the respective Yr gene’s positive controls as identified by linked SNP markers genotyping. Bars of “Combined†represents the frequency of lines containing amplification for all the markers of the respective Yr gene as a haplotype. Markers IWB44454, IWB44619 and T_LYr5 gave ambiguous detection because they amplified the same allele in both positive and negative controls.
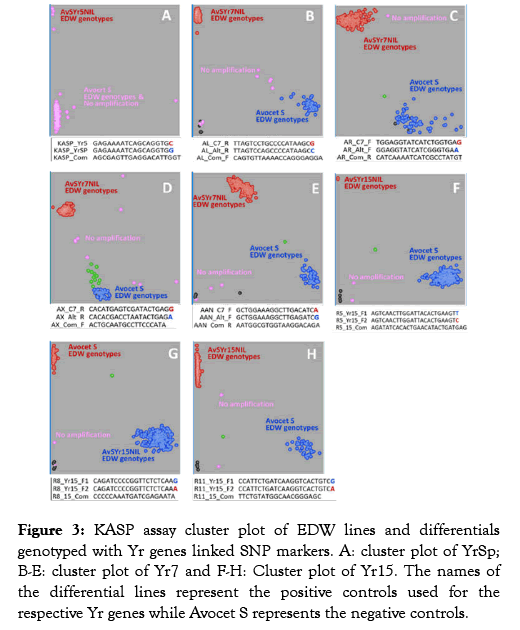
Figure 3:KASP assay cluster plot of EDW lines and differentials genotyped with Yr genes linked SNP markers. A: cluster plot of YrSp; B-E: cluster plot of Yr7 and F-H: Cluster plot of Yr15. The names of the differential lines represent the positive controls used for the respective Yr genes while Avocet S represents the negative controls.
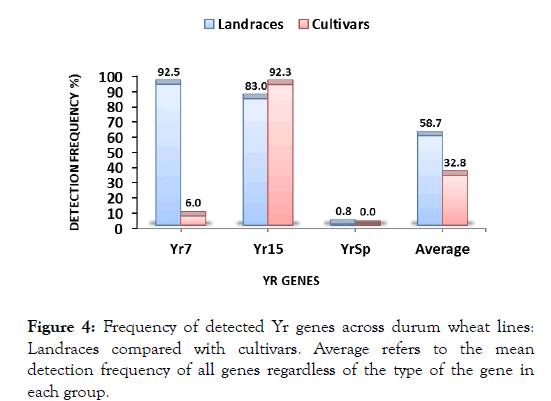
Landraces vs. cultivars and gene combinations
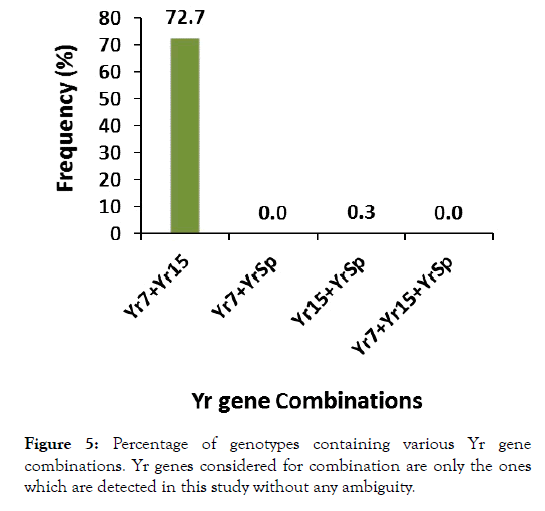
Considering the tested line groups separately, Yr7, Yr15 and YrSp were detected in landraces while only Yr7and Yr15 were in cultivars (Figure 4). Altogether, average detection of these genes was, 58.7%, and 32.8% within landraces and cultivars respectively. When assessing the presence of gene combinations across the lines (Figure 5), 218 of them had a combination of Yr7+Yr15 and just a single line had Yr15+YrSp combination, while none of the lines had the combinations Yr7+YrSp, and Yr7+Yr15+YrSp (Figure 5). Within the 124 phenotypically resistant lines to the three isolates, there was no detection for YrSp while Yr7 and Yr15 were detected in 9 and 19 lines respectively. Of all the resistant lines, only 2 lines exhibited absence of any of the Yr genes considered for detection in the study.
Figure 4:Frequency of detected Yr genes across durum wheat lines: Landraces compared with cultivars. Average refers to the mean detection frequency of all genes regardless of the type of the gene in each group.
Figure 5:Percentage of genotypes containing various Yr gene combinations. Yr genes considered for combination are only the ones which are detected in this study without any ambiguity.
Discussion
Screening/seedling test
High yielding semi-dwarf wheat varieties introduced from the Consultative Group on International Agricultural Research (CGIAR) centers succumb to new races of the rust pathogen soon after their cultivation by farmers. Ethiopian durum landraces serve as useful sources of diversity for important traits that include diseases resistance, drought tolerance, etc. Therefore, 300 durum wheat lines were screened against the three virulent isolates (Pst_Is1, Pst_Is4, Pst_Is8) in search of potential resistance lines. In total, considerably higher number (202 (67.3%)) of the lines were resistant to Pst_Is4 than the other two isolates while 51 (17%) of the lines exhibited the highest susceptibility to Pst_Is8 concluding that most of the lines are well adapted to Pst_Is4 than to Pst_Is1 and Pst_Is8 (from Meraro). It also signifies importance of using various races of varying virulence for screening durum genetic pool and possible gene deployment. In assessing the extent of lines constantly resistant to all the three isolates, the landraces took the larger number (120) than the cultivars (4). This could be because the landraces are grown for a long time in the pathogens environment which has given them a better chance to develop resistance to the pathogen population. Four durum cultivars (Cocorit/71, Yerer, Obsa and Dire) constantly exhibited resistance to all Pst isolates. Though not exclusively on durum, similar studies on identification of resistance sources in wheat have been done by several authors. Wan et al. [19] identified relatively smaller number of resistant accessions (13) in their evaluation of 178 Ethiopian bread wheat lines across two Ethiopian and three USA Pst races. Eight of the lines were cultivars and five were landraces. In their evaluation of Ethiopian wheat against 20 Pst races, Dawit et al. [47] reported two bread wheat cultivars (Wabe and Tuse) were resistant to all 20 races and cultivar Sofumar resistant to most of the races they studied. Through screening of wheat germplasm from Ethiopia several resistant durum landraces and new genes such as Yr53 [48] Yr64, and Yr65 [31] have been identified and transferred to bread wheat backgrounds. These all reports show that landraces particularly of Ethiopian durum wheat germplasm are potential sources of resistance to Pst races of Ethiopian and exotic origin. Similarly, the seedling evaluation in this study has confirmed resistance to the Pst isolates used.
KASP assay based screening for known Yr genes
Knowing that all the three isolates used in this study were avirulent to Yr5, YrSp and Yr15 (Table 1), we anticipated that the lines constantly resistant to all three Pst isolates might harbour at least one of these genes. Subsequently, we did molecular screening of the lines with KASP SNP markers linked to seven Yr genes already reported in literatures including Yr5, YrSp and Yr15. Overall, the molecular screening enabled unambiguous detection of YrSp, Yr7 and Yr15 in various proportions while Yr1, Yr17 and Yr36 were not detected in any of the tested lines (Table 4). Considering the best amplifying linked marker, Yr15 was the most frequently (88.3%) detected gene followed by Yr7 (81.7%) and YrSP with the least (0.7%) detected one. Yr5 is a race specific resistance gene initially identified in a Triticum aestivum ssp. spelta var. album accession [49]. It is mapped on chromosome 2BL [50-54] and is effective against all Pst races identified in the United States [55] although Wellings and McIntosh [56] reported virulence from two Australian Pst isolates. The fact that Yr5 marker T_LYr5 amplified the same allele in both positive and negative controls makes the detection ambiguous. However, the amplification of the positive control allele in 98.6% of the tested lines makes the detection worth considering because none of the isolates in this study were virulent against it as far as the phenotypic screening is concenrned (Table 1). Further assessment on the presence or absence of this gene in Ethiopian durum wheat may help to decide whether or not to use it in wheat breeding program. YrSp, originally identified from ‘ Spaldings Prolific ’ wheat provides resistance to a broad spectrum of Pst races and is mapped on 2BL [57]. Recently, Marchal et al. [40] identified that YrSp is allelic to Yr5 and is a truncated version of Yr5 sharing 99.8% sequence in common. In the present study, marker KASP_YrSp gave amplification in the positive control and only in two of the tested lines. Absence of the virulence to YrSp and lowest detection frequency in this study are good enough evidences to consider it for introgression in Ethiopian wheat materials to increase its frequency. Being a major gene conferring an all stage resistance, Yr7 is mapped on chromosome 7BS [40] and is detected in many (74.0%) of Ethiopian bread wheats cultivars and elite materials [41]. This is a similar result to the detection frequency (81.7% with the best amplifying marker) in our result although ours is on durum wheat lines. It is among the widely used Yr genes by CIMMYT and deployed in many commercial cultivars [58]. In agreement with the result of the virulence survey on Ethiopian Pst races [47], our result indicates that this gene is not effective any more in Ethiopian condition. Frequent usage of bread wheat cultivars with this gene has probably put a selection pressure on the Pst population and has led to the development of new virulent races. Yr15 is among the useful Yr genes originated from and genetically mapped on chromosome 1BS of wild emmer wheat [59]. It has been introgressed in many bread wheat cultivars [60] and known to provide effective resistance against most Pst races worldwide (http:// wheatrust.org/) and major international isolates. Three KASP SNP markers, identified through bulk segregant analysis [39] used in our study gave amplification in considerable number of the durum lines. Although SNP marker R5 gave the lowest detection frequency, markers R8 and R11 have amplified significantly higher number of lines. The latter two markers can be used for marker assisted introgression of the gene and the lines containing the gene can be targeted as potential donors of Yr15.
Yr1, is mapped on chromosome 2AL. SSR markers have been used for haplotyping of this gene [38,41,61] in many wheat materials and various levels of detection frequencies were reported. With the KASP SNP marker IWB81533, no detection was achieved in the present study. On the other hand, the ambiguous detection obtained by markers IWB44454 and IWB44619 could be due to similarity of the genetic background of both positive and negative controls as Avocet S was used for the development of differentials. Earlier studies showed that Yr17 was effective against races of east Africa [62] and under Ethiopian conditions [47] in bread wheat. Its presence in Ethiopian bread wheat cultivars was also confirmed latter using a haplotype analysis of linked SSR markers [41]. In the present study however, the KASP assay-based SNP markers did not show any presence of the gene in the durum lines. Slight sequence difference between bread wheat and durum wheat genomes inevitably exists and that could have worked in favour of Yr17 presence in the hexaploid wheat than in the tetraploid (durum) wheats. Yr36 is a Hight-temperature Adult Plant (HTAP) stripe rust resistance gene mapped on chromosome 6B and closely linked to the Grain protein content locus Gpc-B1 [37]. The Gpc- B1 locus linked SNP marker wMAS000017 amplified only the allele of the negative control indicating the absence of Yr36 among the tested lines.
Conclusions And Future Directions
In this study relatively large number of phenotypically resistant landrace lines instead of cultivars has been identified. Similarly, in the molecular screening the average frequency of resistant genes detected is more in landraces than in the cultivars. This relative richness of landraces as source of resistance over the cultivars can be considered as an opportunity for enhancing potential of wheat for Pst resistance in further improvement programs. Besides, phenotypically resistant lines in which no Yr genes were detected may contain a new gene. These lines can be good starting material towards identification of the gene through mapping, cloning and functional characterization. The identified resistant genes like Yr15 along its diagnostic markers can be used in wheat marker assisted breeding for Pst resistance. Moreover, lines that contain combination of Yr genes are potential targets for starting Yr gene pyramiding for a quick deployment in Ethiopian wheat production. The diagnostic value of markers in diverse genetic backgrounds could sometimes be inconsistent and detections might be ambiguous as in the case of Yr5. Under such conditions, a rigorous reproducibility test should be done to reach at a conclusive result. Aside from the identification of resistance sources, this double screening approach may be considered as useful strategy for relating phenotypically resistant lines with the Yr genes in the absence of enough molecular information about the genes anticipated. Besides, it provides evidence which helps in making decision on whether to use identified resistance genes and the lines in breeding for Yr resistance and gene pyramiding strategies.
Acknowledgements
This research was financially supported by SCPRID project under the initiative of UK Biotechnology and Biological Sciences Research Council (BBSRC); Ethiopian Institute of Agricultural Research (EIAR) and Addis Ababa University (AAU). Ethiopian Biodiversity Institute (EBI) and Ethio Organic Seed Action (EOSA) provided us germplasm, while Kulumsa, Holeta and Debre-Zeit Agricultural Research Centers provided us accesses to greenhouse facilities and experimental fields. We extend our thanks to Dr Mogen Hovemoller (Aarhus University, Denmark) for providing us the Yr differentials.Conflict of Interest
The authors have not declared any conflict of interests.REFERENCES
- CSA. Agricultural Sample Survey 2017/2018 (2010 E.C.). Vol. 1. Report on Area and Production of Major Crops (Private Peasant Holdings, Meher Season). Online Publication. Central Statistics Agency (CSA), Addis Ababa, Ethiopia. 2017.
- Dereje H, Chemeda F. Epidemics of stripe rust (Puccinia striiformis) on common wheat (Triticum aestivum) in the highlands of Bale, South Eastern Ethiopia. Crop Prot 2006;26:1209-1218.
- Wellings CR. Global status of stripe rust: A review of historical and current threats. Euphytica. 2011;179:129-141.
- Wubishet AT, Chemeda FG, Bekele HE. Effects of environment on epidemics of yellow rust (Puccinia striiformis West.) of bread wheat (Triticum aestivum L.) in Bale highlands, South-eastern Ethiopia. Global J Pests Dis Crop Prot 2015;3:96-107.
- Bekele H, Shambel K, Dereje H. Seasonal variations in the occurrence of wheat stripe rust in Bale highlands. Pest Manage. J Ethiopia 2002;6:65-72.
- Badebo A, Bekele E, Bekele B, Hunde B, Degefu M, Tekalign A, et al. Review of two decades of research on diseases of small cereal crops in Ethiopia. Proceedings of the 14th Annual conference of the Plant protection society of Ethiopia (PPSE). 2008.
- Hulluka M, Woldeab G, Andrew Y, Desta R, Badebo A. Wheat Pathology Research in Ethiopia pp. 173–217. In: Wheat Research in Ethiopia: A historical Perspective. Gebre-Mariam J, Tanner DG and Huluka M (Eds). Addis Ababa, IAR/CIMMYT. 1991.
- Badebo A, Stubbs RW, Van Ginkel M, Gebeyehu G. Identification of resistance genes to Puccinia striiformis in seedlings of Ethiopian and CIMMYT bread wheat varieties and lines. Neth J Pl Pathol. 1990;96:199-210
- Solh M, Nazari K, Tadesse W, Wellings CR. The growing threat of stripe rust worldwide Borlaug Global Rust Initiative (BGRI) conference. Beijing, China. 2012.
- Roelfs AP, Singh RP, Saar EE. Rust diseases of wheat: Concepts and methods of disease management, CIMMYT: Mexico, D.F, Mexico.1992;1-81.
- Chen XM. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can J Plant Pathol 2005;27:314-337.
- Chen XM. High-temperature adult-plant resistance, key for sustainable control of stripe rust. Am J Plant Sci 2013;4:608-627.
- Atilaw A, Bishaw Z, Eticha F, Gelalcha S, Tadesse Z. Controlling wheat rusts and ensuring food security through deployment of resistant varieties in Ethiopia. Proceedings of the 2nd International Wheat Stripe Rust Symposium. 2014.
- Belay G, Tesemma T, Becker HC, Merker A. Variation and interrelationships of agronomic traits in Ethiopian tetraploid wheat landraces. Euphytica 1993;71:181-188
- Harlan JR. Ethiopia: A centre of diversity. Econ Bot 1969;23:309-314.
- Harlan JR. Agricultural origins: Centres and non-centres. Science 1971;174:468-473.
- Aklilu M. Wheat rust races identified in virulence survey in Ethiopia. p. 458-461. In: The Ninth Regional Wheat Workshop For Eastern, Central and Southern Africa. Tanner DG, Payne TS, and Abdalla OS (Eds). Addis Ababa, Ethiopia. 1995.
- Dawit W, Flath K, Weber WE, Schumann E, Kosman E. Virulence and diversity of Puccinia striiformis f. sp. tritici in Ethiopia. Can. J. Plant Pathol 2009;31:211-219.
- Wan A, Kebede T, Habtemariam Z, Bekele H. Virulence characterization of wheat stripe fungus Puccinia striiformis f.sp. tritici in Ethiopia and Evaluation of Ethiopian wheat germ-plasm for resistance to races of the pathogen from Ethiopia and the United States. Plant Diseases 2017;101:73-80.
- Hussein S, Pretorius ZA. Leaf and stripe rust resistance among Ethiopian grown wheat varieties and lines. Ethiop J Sci. 2005;28:23-32.
- Vanzetti LS, Campos P, Demichelis M, Lombardo LA, Aurelia PR. Identification of leaf rust resistance genes in Selected Argentinean bread wheat cultivars by gene postulation and molecular markers. Electronic Journal of Biotechnology. 2011;14:9-9.
- Goutam U, Kukreja S, Yadav R, Salaria N, Thakur K, et al. Recent trends and perspectives of molecular markers against fungal diseases in wheat. Front Microbiol 2015;6:861.
- Robert O, Abelard C, Dedryve F. Identification of molecular markers for the detection of the yellow rust resistance gene Yr17 in wheat. Mol Breed. 1995;5:167-175.
- Peng JH, Fahima T, Roeder MS, Huang QY, Dahan A. A high-density molecular map of chromosome region harbouring stripe-rust resistance genes YrH52 and Yr15 derived from wild emmer wheat, Triticum dicoccoides. Genetics. 2000;109:199-210.
- Singh RP, Nelson JC, Sorrels ME. Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 2000;40:1148-1155.
- Helguera M, Khan IA, Kolmer J, Lijavetzky D, Zhong-Qi L. PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Sci. 2003;43:1839-1847.
- Smith P, Koebner R, Boyd L. The development of a STS marker linked to a yellow rust resistance derived from the wheat cultivar Moro. Theor Appl Genet 2002;104:1278-1282.
- Wang L, Ma J, Zhou R, Wang X, Jia J. Molecular tagging of the yellow rust resistance gene Yr10 in common wheat, PI178383 (Triticum aestivum L). Euphytica 2002;124:71-73.
- Asad MA, Xia X, Wang C, He Z. Molecular mapping of stripe rust resistance gene YrSN104 in Chinese wheat line Shaannong 104. Hereditas 2012;149:146-152.
- Liu J, Chang Z, Zhang X, Yang Z, Li X, Jia Ji, et al. Putative Thinopyrum intermedium-derived stripe rust resistance gene Yr50 maps on wheat chromosome arm 4BL. Theor Appl Genet. 2013;126:265-274.
- Cheng P, Xu LS, Wang MN, See DR, Chen XM. Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor Appl Genet 2014;127:2267-2277.
- Randhawa M, Bansal U, Valárik M, Klocová B, Doležel J, Bariana HB. Molecular mapping of stripe rust resistance gene Yr51 in chromosome 4AL of wheat. Theor Appl Genet 2014;127:317-324.
- Zhou XL, Han DJ, Chen XM, Gou HL, Guo SJ, Rong L. Characterization and molecular mapping of stripe rust resistance gene Yr61 in winter wheat cultivar Pindong 34. Theor Appl Genet 2014;127:2349-2358.
- Zhou XL, Wang MN, Chen XM, Lu Y, Kang ZS, Jing JX. Identification of Yr59 conferring high temperature adult plant resistance to stripe rust in wheat germplasm PI178759. Theor Appl Genet. 2014;127:935-945.
- Uauy C, Brevis J, Chen X, Khan I, Jackson L, Chicaiza O. High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor Appl Genet 2005;112:97-105.
- Distelfeld A, Uauy C, Fahima T, Dubcovsky J. Physical map of the wheat high-grain protein content gene Gpc-B1 and development of a high-throughput molecular marker. New Phytol 2006;169:753-763.
- Fu DL, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X. A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 2009;323:1357–1360.
- Bansal UK, Hayden MJ, Keller B, Wellings CR, Park RF, Bariana HS. Relationship between wheat rust resistance genes Yr1 and Sr48 and a microsatellite marker. Plant Pathol 2009;58:1039-1043.
- Ramirez-Gonzalez RH, Segovia V, Bird N, Fenwick P, Holdgate S, Berry S. RNA-seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol J 2015;13:613-624.
- Marchal C, Zhang J, Zhang P, Fenwick P, Steuernagel B, Adamski NM. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nature Plants 2018;4: 662-668.
- Dawit W, Badebo A, Hunde B, Kassa D, Roder MS. Haplotype analysis of Ethiopian bread wheat (Triticum aestivum) cultivars and elite lines for yellow rust resistance genes using linked molecular markers. Afr J Biotechnol 2019;18: 37-57.
- Singh N, Choudhury DR, Singh AK, Kumar S, Srinivasan K. Comparison of SSR and SNP Markers in estimation of genetic diversity and population structure of indian rice varieties. PLoS ONE. 2013;8: e84136.
- Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using competitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed. 2014;33:1-14.
- McNeal FH, Konzak CF, Smith EP, Tate WS, Russell TS. A uniform system for recording and processing cereal research data. U.S. Department, Agric. Res. Serv, ARS. 1971.
- Wheat Training. Wheat DNA extraction in 96 well plate. 2019.
- LGC Genomics: Genotyping services | LGC Biosearch Technologies.
- Dawit W, Flath K, Weber WE, Schumann E, Röder MS, Chen X. Postulation and mapping of seedling stripe rust resistance genes in Ethiopian bread wheat cultivars. Journal of Plant Patholology 2012;94:403-409.
- Xu LS, Wang MN, Cheng P, Kang ZS, Hulbert SH, Chen XM. Molecular mapping of Yr53, a new gene for stripe rust resistance in durum wheat accession PI 480148 and its transfer to common wheat. Theor Appl Genet 2013;126:523-533.
- Macer RCF. The formal and monosomic genetic analysis of stripe rust (Puccinia striiformis) resistance in wheat. Proceedings of the 2nd International Wheat Genetics Symposium. 1966.
- Sun Q, Wei Y, Ni Z, Xie C, Yang T. Microsatellite marker for yellow rust resistance gene Yr5 in wheat introgressed from spelt wheat. Plant Breed. 2002;121:539-541.
- Chen X, Soria MA, Yan G, Sun J, Dubcovsky J. Development of sequence tagged site and cleaved amplified polymorphic sequence markers for wheat stripe rust resistance gene. Crop Sci. 2003;43:2058-2064.
- Yan G, Chen X, Line R, Wellings C. Resistance gene-analog polymorphism markers co-segregating with the Yr5 gene for resistance to wheat stripe rust. Theor Appl Genet. 2003;106:636-643.
- Smith PH, Hadfield J, Hart NJ, Koebner RMD, Boyd LA. STS markers for the wheat yellow rust resistance gene Yr5 suggest a NBS–LRR-type resistance gene cluster. Genome. 2007;50:259-265.
- Naruoka Y, Ando K, Bulli P, Muleta KT, Rynearson S. Identification and validation of SNP markers linked to the stripe rust resistance gene Yr5 in wheat. Crop Sci. 2016;56:3055-3065.
- Wan AM, Chen XM. Virulence characterization of Puccinia striiformis f. sp. tritici using a new set of Yr single-gene line differentials in the United States in 2010. Plant Dis 2014;98:1534-1542.
- Wellings CR, McIntosh RA. Puccinia striiformis f. sp. tritici in Australasia: Pathogenic changes during the first 10 years. Plant Pathol. 1990;39:316-325.
- Feng JY, Wang MN, Chen XM, See DR, Zheng YL, Chao SM, et al. Molecular mapping of YrSP and its relationship with other genes for stripe rust resistance in wheat chromosome 2BL. Phytopathology. 2015;105:1206-1213.
- Van Ginkel M, Rajaram S. Breeding for durable resistance to diseases: An international perspective. In: Durability of Disease Resistance. Jacobs T, Parlevliet JE (Edn) Wageningen, The Netherlands. 1993.
- Sun GL, Fahima T, Korol AB, Turpeinen T, Grama A, et al. Identification of molecular markers linked to the Yr15 stripe rust resistance gene of wheat originated in wild emmer wheat, Triticum dicoccoides. Theor Appl Genet. 1997;95:622-628.
- Yaniv E, Raats D, Ronin Y, Korol AB, Grama A. Evaluation of marker-assisted selection for the stripe rust resistance gene Yr15, introgressed from wild emmer wheat. Molecular Breeding. 2015;35:43.
- Hasancebi S, Mert Z, Ertugrul F, Akan K, Aydin Y. An EST-SSR marker, bu099658, and its potential use in breeding for yellow rust resistance in wheat. Czech Journal of Genetics and Plant Breeding. 2014;50:11-18.
- Badebo A, Stubbs RW. Valuable sources of resistance of wheat to the East African yellow rust isolates. Proceedings of a Regional Workshop for Eastern, Central and Southern Africa. 1995.
Citation: Alemu SK, Badebo A, Tesfaye K, Uauy C (2019) Identification of Stripe Rust Resistance in Ethiopian Durum Wheat by Phenotypic Screening and Kompetitive Allele Specific PCR (KASP) SNP Markers. Plant Pathol Microbiol. 10:483. doi: 10.35248/ 2157-7471.19.10.483
Copyright: © 2019 Alemu SK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.