Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2019) Volume 10, Issue 6
Identification of Novel Potential Causal Agents of Fusarium Wilt of Musa sp. AAB in Southern Mexico
Luis David Maldonado-Bonilla*, Mónica Alicia Calderón-Oropeza, José Luis Villarruel-Ordaz and Ana Claudia Sánchez-EspinosaReceived: 12-Apr-2019 Published: 30-Apr-2019, DOI: 10.35248/2157-7471.10.479
Abstract
A major threat for bananas and plantains production is the Panama Disease or Fusarium Wilt caused by Fusarium oxysporum f. sp. cubense. In order to characterize the causal agents of Fusarium wilt in Mexico, a sampling was performed in symptomatic plantains growing in fields of Oaxaca, a coastal southern state of Mexico. A phylogenetic analysis based on the sequences of TEF 1-α and IGS revealed that three isolates belonged to the Fusarium oxysporum species complex, while two other isolates were identified as members of the Fusarium fujikuroi species complex. Furthermore, isolates from the same complex shared the same ITS2 sequence. Inoculation using spores of each isolate on the roots of Musa sp. AAB cv. Manzano produced wilting symptoms of varying severity, suggesting that the Fusarium wilt might not be caused only by Fusarium oxysporum f. sp. cubense. PCR-based detection of Secreted in Xylem (SIX) genes showed that each Fusarium isolate harbored a unique combination of genes typically found in banana pathogens, which might cause the disease.
Keywords
Breeding; Fusarium oxysporum; Musa sp.; Panama disease; genetic diversity; vascular pathogens; SIX genes
Abbrevations
Foc; Fusarium oxysporum f. sp. cubense; FOSC; Fusarium oxysporum Species Complex; FFSC; Fusarium Fujikuroi Species Complex; TEF 1-α; Translation Elongation Factor 1-α; IGS; Nuclear Ribosomal Intergenic Spacer Region; ITS; rDNA Internal Transcribed Spacer; SIX: Secreted in Xylem.
Introduction
Bananas and plantains (Musa spp.) are a main staple food worldwide. Their production represents an important income source in tropical countries where cultivation of those plants is favored [1]. The tropical and subtropical climate conditions also promotes the establishment of phytopathogens such as Fusarium oxysporum f. sp. cubense (Foc), which is the causal agent of Fusarium wilt or Panama disease [2]. The Panama disease caused by Foc is considered the utmost factor hampering the global banana production. Foc is an asexual ascomycete, belongs to the Fusarium oxysporum species complex (FOSC) that produces macroconidia, microconidia, and the quite resistant chlamydospores, that persist in the soil for years. When such asexual spores germinate, hyphae accede into the plant via the roots. Once inside, the fungus invades xylem vessels and gets dispersed throughout the whole plant, causing yellowing and wilting in old leaves, browning in the vascular tissue and splitting of the pseudostem [3]. Based on the affected Musa host plant, Foc is classified into race 1, 2 and 4 distributed in the eight clonal lineages [3]. Foc race 1 (Foc1) causes disease in the Gros Michel subgroup (genotype AAA), and the Silk subgroup (genotype AAB). Foc1 was responsible of the Panama disease epidemic in the 19th century, that devastated the production of bananas, but it is avirulent to the resistant AAA Cavendish cultivar, whose introduction replenished banana production [3]. Foc race 2 (Foc2) is virulent in the Bluggoe subgroup that carries genotype ABB. Foc race 4 (Foc4) is virulent in Cavendish banana plants (genotype AAA) and cause an enhanced Panama disease in the hosts of Foc1. Because the disease can be present in subtropical or tropical conditions, Foc4 is divided into subtropical race 4 (STR4) and tropical race 4 (TR4) [4]. Since Musa spp. are asexually propagated clones, they have reduced genetic variability, which has promoted the emergence and rapid spreading of the TR4. Panama disease is currently a serious threat for worldwide banana production as TR4 has been detected in South East Asia, Australia, Africa, the Middle East, India, Pakistan, Lebanon [5], and recently in Israel [6]. These findings illustrate how TR4 has been dispersing, thus, identification of causal agents in banana plantations should include the monitoring of TR4 as a necessary early step to prevent economic losses or even export bans. Furthermore, little is known about the diversity of Foc in Latin America. Foc1 isolates have been detected in diverse banana and plantains cultivars in southern Brazil [7], Minas Gerais at the Southeastern Brazil [8], and recently in Puerto Rico [9]. It is worth to mention that a new lineage within Foc was also identified in Goiás, central Brazil, and it is highly virulent in Latundan bananas (AAA) [10]. To our knowledge, there are no reports of TR4 in Latin America, however, further screenings in diseased plants can broaden our knowledge about the fungal biodiversity causing the Panama disease.
States in southern and southeast Mexico with a coastline on the Pacific Ocean are the major producers of bananas and plantains. More than 2000 tons of bananas and plantains are produced per year [11]. Little is known about the incidence of the causal agents of Fusarium wilt and its corresponding genetic diversity in Mexico. In this work, we present the identification of novel pathogenic fungi isolated from plantains (Musa sp. AAB) growing on plantations on the Pacific coast of Oaxaca, a state of the Southeastern Mexico that contributes to the overall local production of plantains. Isolates were classified based on the sequencing of the known markers Translation Elongation Factor 1-α (TEF 1-α) and the Nuclear Ribosomal Intergenic Spacer Region (IGS) [12]. Analysis of the rDNA Internal Transcribed Spacer 2 (ITS2) indicated that each species complex carries at least one sequence variant of this spacer. Foc and related organisms were identified in this search, and its virulence towards Musa sp. AAB was demonstrated. The isolation of fungi that do not belong to the FOSC suggested that Fusarium wilt in plantains is not restricted to Foc. PCR-based amplification of Secreted in Xylem (SIX) genes in these novel isolates corresponds to the pool of SIX genes detected in previously reported Foc1 and Foc2 strains. As in other phytopathogenic F. oxysporum isolates, mechanisms of horizontal transfer of SIX genes might facilitate adaptations required to colonize plantains.
Methods
Isolation of fungi
Two samples from plantations of Musa sp. AAB cv. Macho (Subgroup ‘Plantain’) and Musa sp. AAB cv. Manzano (subgroup Silk) were obtained at the municipality of Villa de Tututepec de Melchor Ocampo, in the region Pacific coast region of Oaxaca, Mexico (16° 08′N, 97° 36′ W). Both cultivars are susceptible to Panama disease [13]. Pseudostems and leaves from plants showing symptoms of wilting were harvested. Tissue samples were washed with sterile water and cut to obtain square-shaped pieces of approximately 0.5 × 0.5 mm. Squares were subjected to a superficial asepsis with 3% sodium hypochlorite for 1 min, and then, were rinsed with sterile distilled water. They were placed in Potato Dextrose Agar (PDA) plates and incubated for 1 week at room temperature. Visible independent mycelia were subsequently transferred into fresh PDA plates until morphologically unique and independent isolates were obtained. The name and source of isolates are shown in Table 1. Isolates showing mycelium reminiscent to the Fusarium genus were analyzed by lactophenol-cotton blue staining and optical microscopy.
| Name | Source | Part of the Plant |
|---|---|---|
| M1a | Musa sp. AAB cv. Macho | Leaf |
| M5 | Musa sp. AAB cv. Manzano | Pseudostem |
| M7 | Musa sp. AAB cv. Macho | Leaf |
| M104 | Musa sp. AAB cv. Manzano | Pseudostem |
| M108 | Musa sp. AAB cv. Manzano | Pseudostem |
Table 1: Fusarium sp. isolates identified in symptomatic plantains in the coast of Oaxaca.
Polymerase chain reaction
Mycelia from the isolates grown in PDA were harvested, frozen in liquid nitrogen and ground with a mortar and pestle. DNA extraction was performed according to a published protocol [14] and the integrity of samples was confirmed by 1% agarose electrophoresis coupled with ethidium bromide staining and UV radiation. DNA from each isolate was diluted and used as template to amplify a fragment of TEF 1-α gene and the complete IGS region according to established PCR conditions [12] by using Recombinant Taq polymerase (Invitrogen). The complete ITS that encompasses the ITS1, 5.8S rDNA and ITS2 was also amplified [15] by using Pfu polymerase (Agilent). The oligonucleotides are enlisted in Table 2.
| Template | Oligonucleotide pairs | Tm (°C) | Amplicon Size (bp) |
|---|---|---|---|
| TEF 1-a | ef1: 5’-ATGGGTAAGGARGACAAGAC-3’ | 53 | ~700 |
| ef2: 5’-GGARGTACCAGTSATCATGTT-3’ | |||
| IGS | iNL11: 5’-AGGCTTCGGCTTAGCGTCTTAG-3’ | 62 | ~2200 |
| iCNS1: 5’-TTTCGCAGTGAGGTCGGCAG-3’ | |||
| ITS | ITS1: 5’-TCCGTAGGTGAACCTGCGG-3’ | 55 | ~540 |
| ITS4: 5’- TCCTCCGCTTATTGATATGC -3’ | |||
| SIX1a | SIX1F: 5’-CCCTCTCAATCCTTGGGTTT-3’ | 58 | 153 |
| SIX1R: 5’-TAGTGTCATTCCACGGCAAA-3’ | |||
| SIX6 | SIX6F: 5’-GACMTATGACCGCTCCGTYTG-3’ | 58 | 197 |
| SIX6R: 5’-GGGWMGTTTTCCACGAGACAAG-3’ | |||
| SIX9 | SIX9F: 5’-CTTCTCCCGAAGCTTCTCCT-3’ | 58 | 164 |
| SIX9R: 5’-TTGGAAGCCCAGTTGTAAGG-3’ | |||
| SIX13 | SIX13F: 5’-CGATGGAGTAAATGGGGAAA-3’ | 58 | 196 |
| SIX13R: 5’-TTGTAAACTGTCCCGTGCTG-3’ |
Table 2: List of oligonucleotide sequences used on this study.
Cloning of TEF1-α and IGS
Once the PCR products of TEF 1-α and IGS were confirmed by electrophoresis, the amplicons were cloned into pGEM T-Easy (Promega) by following manufacturer’s protocol. Chemically competent E. coli DH10B cells were transformed with the ligation reactions. Positive transformed cells were selected in LB plates supplemented with 100 μg /mL ampicillin and 40 μL of 50mg/mL X-Gal. Cells carrying the expected plasmids were grown overnight in LB medium supplemented with 100 μg/mL ampicillin. Plasmid purification was performed using the alkaline lysis method, and successful cloning was confirmed by restriction digestion with EcoRI.
DNA sequencing
DNA was sequenced by capillary electrophoresis at Macrogen Inc, Korea (https://dna.macrogen.com/eng/). Plasmids carrying the TEF 1-α and IGS clones were sequenced using the T7 and SP6 universal primers. Primer walking was necessary to obtain the complete sequences of IGS. The complete ITS PCR products were sequenced by using the oligonucleotides used for amplification. Elecropherograms were analyzed in Chromas Version 2.6.5, and sequences were deposited in GeneBank (https://www.ncbi.nlm. nih.gov/nucleotide/). List of the accession numbers of TEF 1-α and IGS are included in the Table 3. The ITS sequences are deposited in GeneBank with the accession numbers MK250065 to MK250069.
| ID/NRRL | Host | TEF | IGS |
|---|---|---|---|
| M1a | Musa sp. AAB Macho | MG018806 | MG193546 |
| M5 | Musa sp. AAB Manzano | MG018805 | MG193545 |
| M7 | Musa sp. AAB Macho | MG018802 | MG193542 |
| M104 | Musa sp. AAB Manzano | MG018803 | MG193543 |
| M108 | Musa sp. AAB Manzano | MG018804 | MG193544 |
| NRRL5883 | Wheat | AF107883.1, FD_00001_EF-1a | FD_00001_rDNA-IGS |
| NRRL22172 | Maize | AF160262.1, | FD_01185_IGS-2733 |
| VI01096 | Barley | AJ543567.1 | AY250995.1 |
| VI01087 | Bread wheat | AJ543570.1 | AY253668.1 |
| NRRL13999 | Sugarcane | AF160278.1 | FD_01170_IGS-2733 |
| NRRL22016 | Corn | AF160289.1 | FD_01160_IGS-2733 |
| NRRL25226 | Mango | AF160281 | GU737449.1 |
| NRRL47473 | Mango | GU737416.1 | GU737473.1 |
| NRRL36266 | Cyclamen | FJ985339.1 | FJ985572.1 |
| NRRL26033 | Tomato | AF008507.1 | FJ985484.1 |
| NRRL26203 | Tomato | AF008501.1 | FJ985487.1 |
| NRRL25603 | Musa sp. AAA Cavendish | AF008487.1 | FJ985480.1 |
| NRRL25607 | Musa sp. ABB Bluggoe | AF008489.1 | FJ985469.1 |
| NRRL25609 | Musa sp. ABB Harare | AF008490.1 | FJ985481.1 |
| NRRL26022 | Musa sp. ABB Pisang Awak | AF008491.1 | FJ985482.1 |
| NRRL26029 | Musa sp. AAB Silk | AF008493.1 | FJ985483.1 |
| NRRL36114 | Musa sp. ABB Pisang Manurung | FJ985328.1 | FJ985561.1 |
| NRRL36118 | Musa sp. ABB Pisang Awak | FJ985330.1 | FJ985563.1 |
| NRRL31649 | Musa sp. | FD_01770_EF-1a | FD_01770_IGS-2733 |
| TR2 | Musa sp. ABB Bluggoe | KC889020.1 | KC869389.1 |
| NRRL26406 | Muskmelon | AF008504.1 | FD_01193_EF-IGS |
| NRRL26960 | Carnation | AF246839.1 | FJ985514.1 |
| NRRL38302 | Pinus | GU170559.1 | FJ985679.1 |
| NRRL25375 | Human | AY527521.1 | AY527718.1 |
Table 3: List of sequences of TEF 1-a and IGS used to phylogenetic.
Alignment and phylogenetic analysis:
Sequences of TEF 1-α and IGS from the isolates reported here, and reference sequences retrieved form GeneBank and Fusarium- ID [16] (ESM1) were aligned separately by using the ClustalW algorithm. Phylogeny of concatenated alignments were subject to Maximum Likelihood (ML) method with 1000 bootstrap replicates by using PhyML 3.0 [17]. The phylogenetic tree was built in MEGA6 [18]. The Clustal W algorithm was also used to align the ITS2 sequences of all isolates together with previously reported sequences of F. oxysporum NRRL22902 (U34566.1), F. subglutinans NRRL22034 (GQ167235.1). These reference sequences displayed 100% identity with the sequences of the isolates. The ITS2 of F. fujikuroi NRRL13566 (AY249382.1) was included as reference of ITS2 Type II [15].
In vitro propagation of plants
Micropropagation of Musa sp. AAB cv. Manzano plants was established by following reported procedures [19,20]. Briefly, axenic plants were produced after surface disinfection of flower buds incubated in MS medium [21] supplemented with 5mg/L Benzyl amino purine (BAP) and 5 g/L Phytagel as gelling agent. Direct organogenesis was induced after 8 weeks of incubation. The shoots were then transferred into MS medium supplemented with 0.5 mg/L BAP and Phytagel. Newly emerging shoots were excised and transferred into solid MS supplemented with 0.1 mg/L of Indole-3-acetic acid (IAA) to induce rooting. After 4 weeks, rooted plantlets were transferred into plastic containers with vermiculite rinsed with 25% MS solution without sucrose. Under these conditions, plants were grown and acclimatized until reach a 6-leaf stage, then, they are harvested and used for infection experiments. All procedures of plant tissue culture were run in a growth chamber at 26°C with a 16 h light and 8 h darkness photoperiod.
Pathogenicity assays
The experimental design was based on the aggressiveness test previously reported [10], it was performed twice. Healthy plants generated as described above were harvested, and their roots were thoroughly washed with distilled water. Every Fusarium isolate was grown in PDA at 28°C for 1 week. Spores were then collected with sterile distilled water and adjusted to a concentration of 106 spores/ml. Inoculation with every Fusarium isolate was performed by submerging the roots of five independent plants in the spore solutions for 30 seconds. After inoculations, plants were planted in plastic bags containing a mixture of peat:vermiculite 1:1. Symptom development and severity in the leaves was monitored and scored based on previous reported criteria [22].
Detection of SIX genes
DNA from fungal isolates was used as a template to amplifying SIX genes commonly present in Foc [23]. Oligonucleotides were designed according to SIX gene sequences available in GeneBank (Table 2). PCR was performed by using the qARTA Taq polymerase following the manufacturer’s recommendations. All PCR reactions started with a 5 min denaturation at 95°C step, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. After amplification, reactions were analyzed by electrophoresis in 2.5% agarose gels stained with ethidium bromide under UV illumination. Amplification of the TEF 1-α was used as a positive control.
Results
Fusarium isolates from the coast of Oaxaca belong to two distinct species complexes
Inspection of fungal diversity coming from symptomatic plants resulted in the isolation of five independent morphotypes (Table 1). Phylogenetic analysis of the markers TEF 1-α and IGS was performed to examine the genetic diversity of such isolates (Figure 1). A ML tree revealed that isolates M1a and M5 were closely related to Foc NRRL25609, an isolate from Musa sp. ABB classified as Foc2. It belonged to the clonal lineage VIII and vegetative compatibility group (VCG) 01214 [24,25]. In accordance with this finding, most of the isolates recently identified in Puerto Rico also belonged to the VCG 0124 [9]. The isolate M104 was not related to the known races of Foc, instead, it was more related to the pathogens of tomato NRRL26033 and NRRL26203 [26]. All members of FOSC in this analysis are clustered together, including F. foetens, which were included as a reference of another member of FOSC. This phylogenetic analysis revealed that F. sacchari and F. verticillioides were species assigned to M7 and M108, respectively. Both are members of the F. fujikuroi species complex (FFSC). M7 is quite similar to NRRL31649, also isolated form Musa sp., and F. sacchari NRRL13999 isolated from sugar cane. The phylogenetic analysis of M108 reveals that F. verticillioides NRRL22172 is the most related species. Due to evident polymorphisms in the sequences of ITS2, that intervenes the 5.8S and 18S ribosomal genes, the ITS2 sequences are grouped into two divergent types in the Fusarium genus. Members of FOSC primarily carry variants of the ITS2 Type I, although some members FFSC have also copies of this Type, which might be consequence of interspecies hybridization or gene duplication events [15]. The complete ITS of the isolates was sequenced and aligned together with reference sequences of ITS2 Type I and Type II. All the isolates presented here carried at least one copy of the ITS2 Type I (Figure 2). It is worth to mention that none of the identified isolates was related to the TR4. Phylogenetic analysis presented here is supported by lactophenol cotton blue staining and optical microscopy observations, which revealed the production of characteristic structures such as chlamydospores and false heads in the isolates from FOSC. The FFSC isolate M7 produced septate mesoconidia, the isolate M108 produced chains of microconidia reminiscent of F. verticillioides (Supplementary Information). Taken together, morphology and phylogenetic analysis illustrate that fungi from FOSC and FFSC were found in the organs of plantains displaying symptoms of Fusarium wilt.
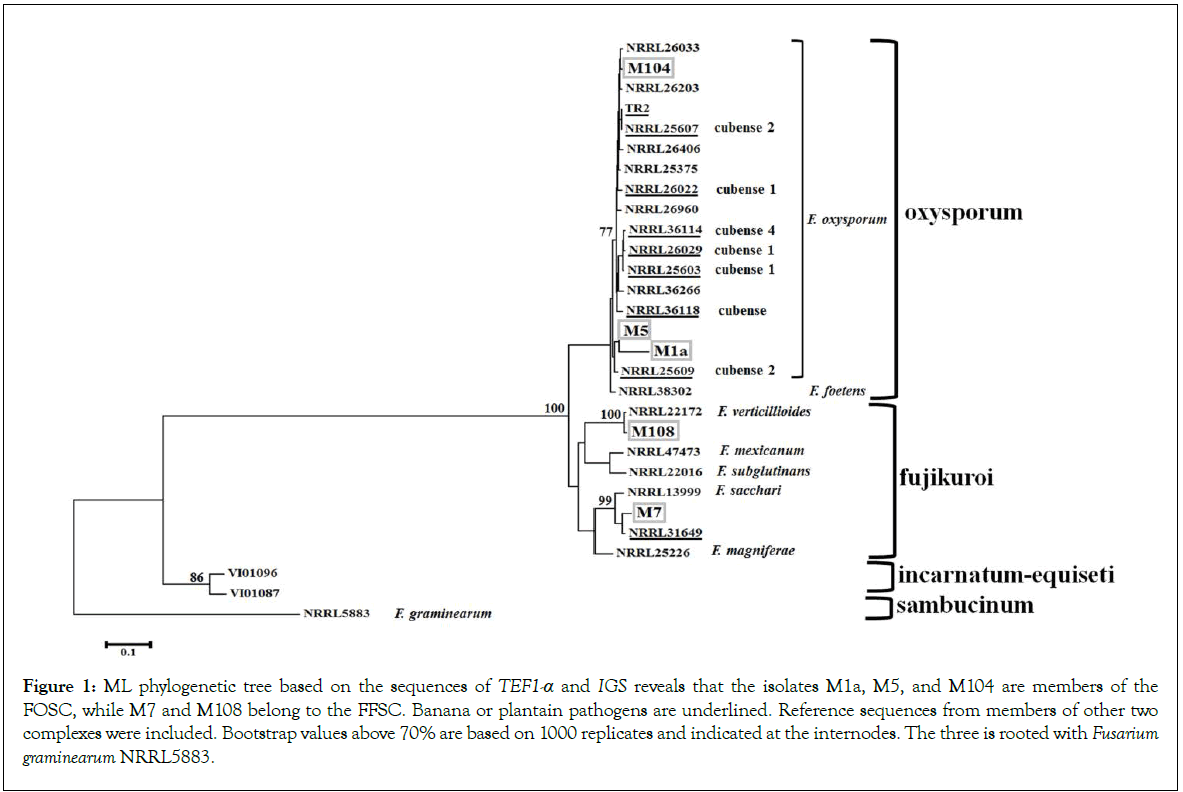
Figure 1: ML phylogenetic tree based on the sequences of TEF1-a and IGS reveals that the isolates M1a, M5, and M104 are members of the FOSC, while M7 and M108 belong to the FFSC. Banana or plantain pathogens are underlined. Reference sequences from members of other two complexes were included. Bootstrap values above 70% are based on 1000 replicates and indicated at the internodes. The three is rooted with Fusarium graminearum NRRL5883.
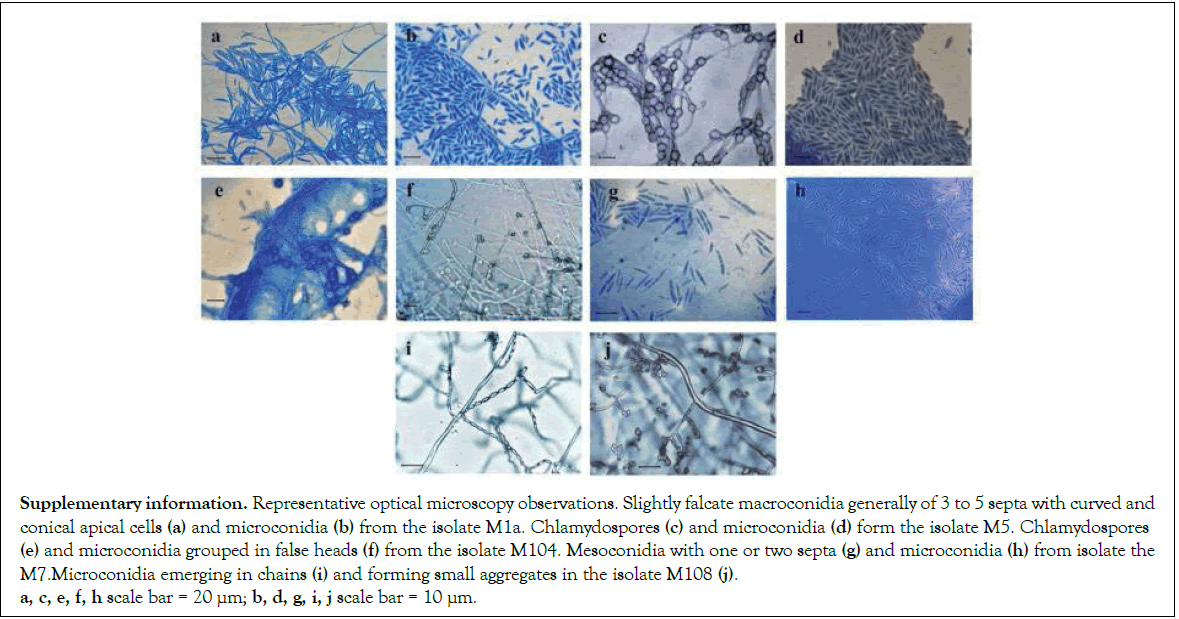
Supplementary information. Representative optical microscopy observations. Slightly falcate macroconidia generally of 3 to 5 septa with curved and conical apical cells (a) and microconidia (b) from the isolate M1a. Chlamydospores (c) and microconidia (d) form the isolate M5. Chlamydospores (e) and microconidia grouped in false heads (f) from the isolate M104. Mesoconidia with one or two septa (g) and microconidia (h) from isolate the M7.Microconidia emerging in chains (i) and forming small aggregates in the isolate M108 (j). a, c, e, f, h scale bar = 20 µm; b, d, g, i, j scale bar = 10 µm.
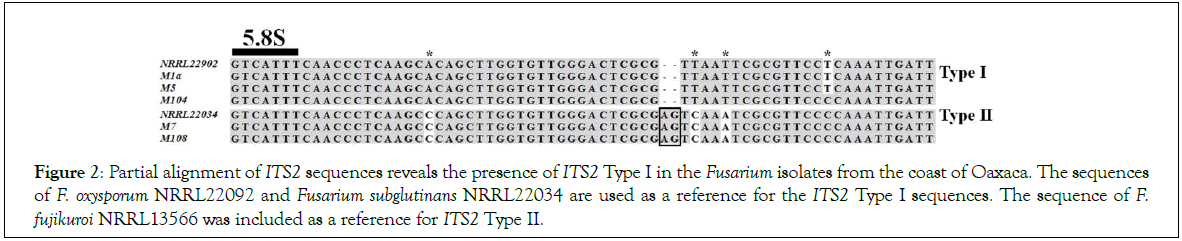
Figure 2:Partial alignment of ITS2 sequences reveals the presence of ITS2 Type I in the Fusarium isolates from the coast of Oaxaca. The sequences of F. oxysporum NRRL22092 and Fusarium subglutinans NRRL22034 are used as a reference for the ITS2 Type I sequences. The sequence of F. fujikuroi NRRL13566 was included as a reference for ITS2 Type II.
Fusarium isolates are virulent in Musa sp. AAB cv. Manzano
A pathogenicity assay was performed in order to confirm whether the identified Fusarium spp. caused symptoms related to Panama disease. Acclimatized plantains obtained by a micropropagation method were infected by submerging their roots in a spore solution. Five plants were used per each treatment, and the experiment was performed twice. Every isolate caused symptoms both in leaves and corms of inoculated plants (Figure 3a); however, the emergence of wilting was dependent on the isolate, and the M5 was one of the FOSC that produced symptoms in leaves sooner, and provoked more severe disease by the end of the experiments (Figure 3b). Yellowing and necrosis caused by isolates of FFSC were weaker, and leaf deformation was detectable.
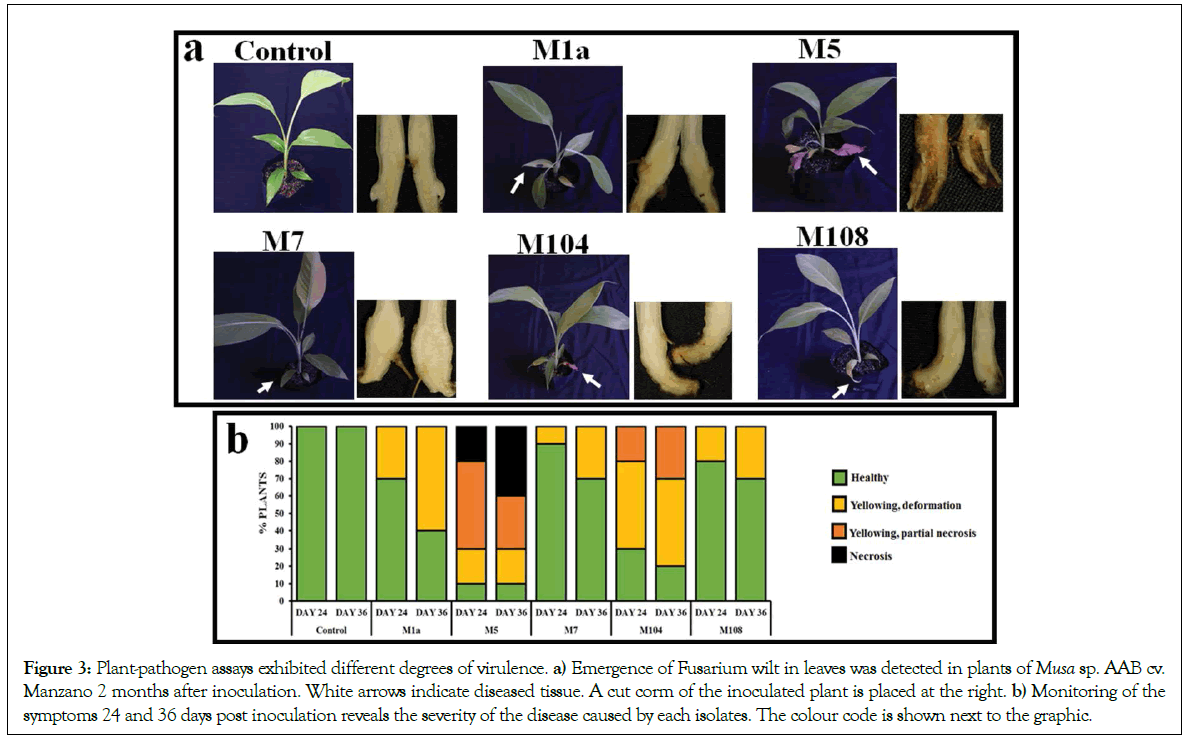
Figure 3:Plant-pathogen assays exhibited different degrees of virulence. a) Emergence of Fusarium wilt in leaves was detected in plants of Musa sp. AAB cv. Manzano 2 months after inoculation. White arrows indicate diseased tissue. A cut corm of the inoculated plant is placed at the right. b) Monitoring of the symptoms 24 and 36 days post inoculation reveals the severity of the disease caused by each isolates. The colour code is shown next to the graphic.
Amplification of SIX genes commonly found in the causal agents of Panama disease
A critical event during colonization the xylem vessels and establishment of the Fusarium wilt is the secretion of small and cysteine-rich proteins known as ‘Secreted in Xylem’ (SIX), which were initially identified in the sap of tomato plants infected with F. oxysporum f. sp. lycopersici (Fol) [22]. So far, there have been 14 SIX genes identified in F. oxysporum. Nevertheless, every forma specialis has a specific combination of such genes, which contributes to disease establishment across a narrow host range [27]. SIX1a, SIX6, SIX9 and SIX13 are often found in isolates of the three Foc races. Additional copies of SIX1 (SIX1b and SIX1c), SIX2 and SIX8 are specific to the race 4 [23]. To get initial molecular evidences regarding the pathogenicity of these isolates, oligonucleotides were designed to amplify the known SIX genes present in Foc. The amplification products are presented in the Figure 4. The SIX1a amplicon was detected in all the isolates regardless the species complex to belong. The SIX6 amplicon is detected in M1a, M5, M104 and M108. SIX9 was detected in isolate M104 and M108, and SIX13 was amplified in isolates M1a, M5 and M104, which were classified as F. oxysporum. M1a and M5 are phylogenetically related, and shared the same combination of the SIX amplicons. However, M5 was the most virulent in our conditions; thus, virulence did not correlate with the presence or lack of these genes. Despite the SIX genes are found in virulent isolates of F. oxysporum, we did amplify selected SIX genes in isolates M7 and M108, both of them belong to the FFSC. We also designed oligonucleotides to amplify SIX2 and SIX8, which are typically detected in the race 4. However, there was no amplification detected in any isolate examined in this research (data not shown).
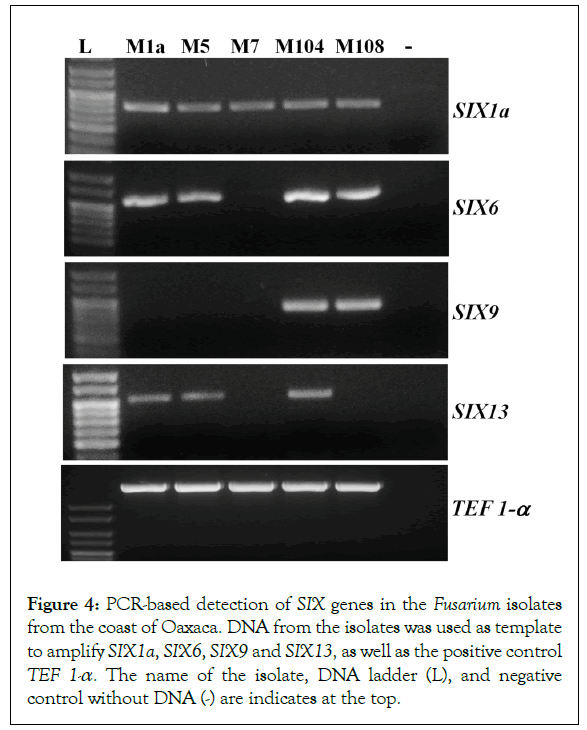
Figure 4:PCR-based detection of SIX genes in the Fusarium isolates from the coast of Oaxaca. DNA from the isolates was used as template to amplify SIX1a, SIX6, SIX9 and SIX13, as well as the positive control TEF 1-a. The name of the isolate, DNA ladder (L), and negative control without DNA (-) are indicates at the top.
Discussion
The FOSC encompasses saprophyte constituents of the rhizosphere and soil borne phytopathogenic fungi, including Foc [28]. The lack of sexual reproduction in F. oxysporum might be compensated by anastomosis which enables horizontal gene transfer (HGT) processes, that could originate over 70 formae especiales distributed in many clonal lineages [29]. In the case of Foc, 23 clonal lineages have been reported, they could emerge from events of HGT events that carried over the genetic information required to parasitize Musa spp.
Although there are reports of Foc in Latin America, we barely know about outbreaks of this pathogen on plantations in Mexico. In order to investigate the genetic diversity of the causal agents of Panama disease, we sampled symptomatic plantains on the coast of Oaxaca, Mexico. After this Focused analysis, we demonstrated that the isolates M1a and M5 are two Foc individuals related to NRRL25609, a reference Foc2 strain isolated from Musa sp. ABB. In this context, the genome B of plantains such as Musa sp. cv. Manzano might encode for factors that facilitate the colonization of Foc2. In spite of the close phylogenetic relationship between M5 and M1a, wilting symptoms produced by both isolates are contrasting, being the M5 the most virulent. Variations in the content and regulation of virulence genes might explain the difference in the symptoms caused by M1a and M5. The ML phylogeny of concatenated TEF 1-α and IGS inferred a close relationship between the virulent isolate M104 and the tomato pathogens NRRL26033, NRRL26023, that belong to a lineage unrelated to Foc [24]. Recently, a wide analysis of the diversity of Foc in Indonesia –the centre of origin of bananademonstrated that Fusarium wilt is not limited to FOSC, since isolates form FFSC and other species complexes are virulent against Gros Michel and Cavendish [30]. F. verticillioides is a well-known pathogen of maize, but some isolates have been identified in rotten banana fruits produced in Mexico [31]. In this work, we identified isolates F. verticillioides M108 and F. sacchari M7, both of which are members of the FFSC. Leaf symptoms caused by the isolates of M108 and M7 are minimal, but it demonstrates that individuals of the FFSC are part of the diversity of Fusarium in plantains AAB in the coast of Oaxaca, Mexico. Their presence in symptomatic plants might be facilitated by virulent organisms such as Foc that suppress the plant immune system to permit the growth of weak pathogens. Fusarium sacchari was also recovered form symptomatic banana plants in Puerto Rico [9], but no symptoms were produced.
Whole genome sequencing of the F. oxysporum model strains has revealed the presence of core and lineage specific (LS) chromosomes [32,33]. LS chromosomes are also known as conditionally dispensable (CD) chromosomes or pathogenicity chromosomes, they are unique to each virulent F. oxysporum isolate and were acquired by HGT [34]. LS chromosomes carry genes encoding virulence factors such as the SIX genes, their own transcriptional regulators and the majority of transposable elements (TE) in the whole genome [33,35,36]. Each LS chromosome was likely outcome from “copy-paste” or “cut-paste” mechanisms promoted by the TE and transposases encoding within [34,35]. After DNA rearrangements, specific combinations of SIX and other genes are formed, and they provide the ability to colonize the roots and suppress the plant’s immune response. The natural selection of every fungus was probably facilitated when the genetic information carried on the LS chromosomes conferred an adaptive advantage. Specific combination of SIX genes might have a more critical role during the invasion of banana plants. For example, homologs of SIX1 are frequently identified in Foc isolates, suggesting that homologs of SIX1 protein play a prominent role in causing disease, regardless the race, plant host and geographic origin [8,37]. Homologs of SIX1 are early expressed in planta both in Foc1 and Foc4 [38], furthermore, the deletion of SIX1a compromises the virulence of TR4 towards Cavendish bananas [39]. SIX1a might target a critical pathway conserved in plants, whose disturbance is necessary to set the Fusarium wilt, since deletion of SIX1 also attenuates the virulence of Fol and F. oxysporum f.sp. conglutinans [22,40]. In accordance with the reports mentioned above, PCR analyses presented here showed that all the isolates possess a copy of SIX1a. The SIX6 gene is also detected in isolates from the coast of Oaxaca. Analysis if SIX6 of Fol suggests a role in suppressing cell death [41] and it might have the same role in Musa pathogens. SIX9 was detected in the isolates M104 and M108, and it was not detected in the isolate M5, for which inoculation displayed the more severe symptoms. The SIX9 gene has been detected in various isolates of Foc1, Foc2 and Foc4 and other formae especiales [23]. However, its role is unknown. The phylogenetic analysis mentioned above revealed that M1a, M5 and M104 are members of FOSC, M1a and M5 are closely related, but M104 derives from a different lineage. This observation might imply that M104 acquired its virulence via an HGT mechanism in a manner unrelated that of M1a and M5. Sequencing of SIX genes in our isolates will be necessary to elucidate the basis of their virulence. SIX13 is found in pathogens of tomato, cucurbits and banana [27]. In our case it was exclusively amplified in members of the FOSC.
SIX genes have been also identified in species from FFSC that infect flower bulbs, such as F. proliferatum and F. agapanthi [42]. Those SIX genes might be acquired by inter-species HGT from FOSC to FFSC [42]. The amplicons of SIX genes in the isolates M7 and M108 presented in this work are hints of HGT. This potential mechanism might have led to the selection of novel virulent strains, or at least, it might have permitted endophytic growth of rhizosphere inhabitant fungi. The potential interspecies hybridization that led the conservation of nonorthologous copies of ITS2 suggests the possibility of the interspecies acquisition of virulence factors, that shape the high genetic diversity of virulent isolates, such as those in Indonesia [30].
Conclusion
Taken together, this work contributes in the identification of novel FOSC and FFSC isolates from the coast of Oaxaca that displayed differential aggressiveness towards Musa sp. AAB cv. Manzano. The ability to cause wilting symptoms and the potential translocation into the plant might be facilitated by the profile of SIX genes. Therefore, we consider the isolates identified here are candidates for Next Generation Sequencing in order to decipher the complete arsenal of virulence factors, the biogenesis and structure of potential LS chromosomes as well as their transfer among Fusarium populations.
Acknowledgments
This research was financially supported by the Universidad del Mar (2IG1801) and Programa Para El Desarrollo Profesional Docente (Grants CA-19 and CA-24). LDMB thanks to the Cátedras CONACYT Program, Grant No. 538. We thank to Edmundo Rosique-Gil and numerous undergraduate students for their valuable technical assistance.
REFERENCES
- http://www.fao.org/3/a-i7410e.pdf
- Stover R. Fusarial Wilt (Panama Disease) of bananas and other Musa species. Commonwealth Mycological Inst. Kew, UK: Commonwealth Mycol Insti 1962.
- Ploetz RC. Fusarium wilt of banana. Phytopatholo 2015;105:1512-1521.
- Fourie G, Steenkamp ET, Ploetz RC, Gordon TR, Viljoen A. Current status of the taxonomic position of Fusarium oxysporum formae specialis cubense within the Fusarium oxysporum complex. Infect Genet Evol 2011;11: 533-542.
- Ordonez N, Seidl MF, Waalwijk C, Drenth A, Kilian A, Thomma BPHJ, et al. Worse comes to worst: Bananas and Panama Disease—when plant and pathogen clones meet. PLoS Pathog 2015;11:e1005197.
- Maymon M, Shpatz U, Harel Y, Levy E, Elkind G, Teverovsky E, et al. First Report of Fusarium oxysporum f. sp. cubense Tropical Race 4 Causing Fusarium Wilt of Cavendish Bananas in Israel. Plant Dis 2018;102:2655.
- Cunha CMS, Hinz RH, Pereira A, Tcacenco FA, Stadnik MJ. Aggressiveness and genetic diversity of Fusarium oxysporum f. sp. cubense from Santa Catarina, Southern Brazil. Trop Plant Pathol 2015;40:326-334.
- Deltour P, França SC, Heyman L, Pereira OL, Höfte M. Comparative analysis of pathogenic and nonpathogenic Fusarium oxysporum populations associated with banana on a farm in Minas Gerais, Brazil. Plant Pathol 2018;67:707-718.
- Garcia RO, Rivera-Vargas LI, Ploetz R, Correll JC, Irish BM. Characterization of Fusarium spp. isolates recovered from bananas (Musa spp.) affected by Fusarium wilt in Puerto Rico. Eur J Plant Pathol 2018;152:599-611.
- Araújo NAF, Pasqual M, Pio LAS, Alves E, De Matos Moura N, Da Costa S. Identification and aggressiveness of four isolates of Fusarium oxysporum f.sp. cubense from Latundan banana in Brazil. J Phytopathol 2017;165: 257-264.
- http://online.pubhtml5.com/clsi/ibhs/
- O’Donnell KO, Gueidan C, Sink S, Johnston PR, Crous PW, Glenn A, et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet Biol 2009;46: 936-948.
- Nadal-Medina R, Manzo-Sánchez G, Orozco-Romero J, Orozco-Santos M, Guzmán-González S. Diversidad genética de bananos y plátanos (Musa spp.) determinada mediante marcadores RAPD. Rev Fitotec Mex 2009;32:1-7.
- Lin YH, Chang JY, Liu ET, Chao CP, Huang JW, Chang PF, et al. Development of a molecular marker for specific detection of Fusarium oxysporum f. sp. cubense race 4. Eur J Plant Pathol (009;123:353-365.
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 1997;7:03-116.
- Geiser DM, Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Zhang N, et al. Fusarium-ID v.1.0: A DNA sequence database for identifying Fusarium. Eur J Plant Pathol 2004;110: 473-479.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0 performance. Syst Biol 2010;59:307-321.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics Analysis version 6.0. Mol Biol Evol 2013;30:2725-2729.
- Uzcátegui JP, Hernández Y, Osorio D, Rivas M. Evaluación del comportamiento in vitro de ápices de plátano Musa AAB cv.Hartón y Hartón Doble Tallo. Prod Agropecu 2010;3:7-12.
- Rios G, Añez N, Ramírez M, Bracho B, Araujo D, Suárez H, et al. In vitro culture of buds, treated with benzyladenine, coming from whole and sectioned corms of plantain Cambur Manzano. Bioagro 2013;25:137-142.
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 1962;15:81-84.
- Rep M, Van Der Does HC, Meijer M, Van Wijk R, Houterman PM, Dekker HL, et al. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 2004;53:1373-1383.
- Czislowski E, Fraser-Smith S, Zander M, O’Neill WT, Meldrum RA, Tran-Nguyen LTT, et al. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol Plant Pathol 2018;19:1155-1171.
- Fourie G, Steenkamp ET, Gordon TR, Viljoen A. Evolutionary relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Appl Environ Microbiol 2009;75:4770-4781.
- Dita MA, Waalwijk C, Buddenhagen I, Souza M, Kema G. A molecular diagnostic for tropical race 4 of the banana Fusarium wilt pathogen . Plant Pathol 2010;348-357.
- Lievens B, Van Baarlen P, Verreth C, Van Kerckhove S, Rep M, Thomma BP. Evolutionary relationships between Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. radicis-lycopersici isolates inferred from mating type, elongation factor-1a and exopolygalacturonase sequences. Mycol Res 2009;113:1181-1191.
- Van Dam P, Fokkens L, Schmidt SM, Linmans JHJ, Corby Kistler H, Ma LJ, et al. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ Microbiol 2016;18:4087-4102.
- Fravel D, Alabouvette C. Fusarium oxysporum and its biocontrol. New Phytol 2003;157:493-502.
- Lievens B, Rep M, Thomma B. Recent developments in the molecular discrimination of formae speciales of Fusarium oxysporum. Pest Manag Sci 2008;64:781-788.
- Maryani N, Lombard L, Poerba YS, Subandiyah S, Crous PW, Kema GHJ. Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the Indonesian centre of origin. Stud Mycol 2019;92:155-194.
- Hirata T, Kimishima E, Aoki T, Niremberg H, O’Donnell K. Morphological and molecular characterization of Fusarium verticillioides from rotten banana imported into Japan. Mycosciene 2001;42:155-166.
- Ma L, Van Der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ Di Pietro A, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nat 2010;464;367-373.
- Van Dam P, Fokkens L, Ayukawa Y, Van Der Gragt M, Ter Horst A, Balázs Brankovics, et al. A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci Rep 2017;7:1-15.
- Vlaardingerbroek I, Beerens B, Schmidt SM, Cornelissen BJC, Rep M. Dispensable chromosomes in Fusarium oxysporum f. sp lycopersici. Mol Plant Pathol 2016;17:1455-1466.
- Schmidt SM, Houterman PM, Schreiver I, Ma L, Amyotte S, Biju Chellappan, et al. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom 2013;14:119.
- Van der Does HC, Fokkens L, Yang A, Schmidt SM, Langereis L, Lukasiewicz JM, et al. Transcription Factors Encoded on Core and Accessory Chromosomes of Fusarium oxysporum Induce Expression of Effector Genes. PLoS Genet 2016;12:e1006401.
- Meldrum RA, Fraser-Smith S, Tran-Nguyen LTT, Daly AM, Aitken EAB. Presence of putative pathogenicity genes in isolates of Fusarium oxysporum f. sp. cubense from Australia. Australas Plant Pathol 2012;41:551-557.
- Guo L, Han L, Yang L, Zeng H, Fan D, Zhu Y, et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f . sp . cubense causing banana vascular wilt disease. PLoS One 2014;9:e95543.
- Widinugraheni S, Niño-Sánchez J, Van Der Does HC, Van Dam P, Garcia-Bastidas F, Subandiyah S, et al A SIX1 homolog in Fusarium oxysporum f . sp. cubense tropical race 4 contributes to virulence towards Cavendish banana. PLoS One 2018;13:e0205896.
- Li E, Wang G, Xiao J, Ling J, Yang Y, Yang Y, et al. A SIX1 homolog in Fusarium oxysporum f. sp. conglutinans is required for full virulence on cabbage. PLoS One 2016;11:e0152273.
- Gawehns F, Houterman PM, Ichou FA, Michielse CB, Hijdra M, Cornelissen BJ, et al. The Fusarium oxysporum Effector SIX6 Contributes to Virulence and Suppresses I-2-Mediated Cell Death. Mol Plant Microbe In 2014;27:336-348.
- Van Dam P, Rep M. The distribution of miniature impala elements and SIX genes in the Fusarium Genus is suggestive of horizontal gene transfer. J Mol Evol 2017;85:14-25.
Citation: Maldonado-Bonilla LD, Calderón-Oropeza MA, Villarruel-Ordaz JL, Sánchez-Espinosa AC (2019) Identification of Novel Potential Causal Agents of Fusarium Wilt of Musa sp. AAB in Southern Mexico. Plant Pathol Microbiol. 10: 479. doi: 10.35248/2157-7471.10.479
Copyright: © 2019 Maldonado-Bonilla LD, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

