Indexed In
- Open J Gate
- Genamics JournalSeek
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2024) Volume 14, Issue 4
Genotype-phenotype Correlations in Inherited Cardiomyopathies, their Role in Clinical Decision-making and Implications in Personalized Cardiac Medicine in Multi-omics as Well as Disease Modelling Eras
Yaqob Samir Taleb1,4*, Paras Memon3, Aftab Jalbani3, Nouf Al-Mutairi2,7, Sarah Al-Mukhaylid2,7, Nawaf Al-Anazi4,6, Abdulkareem Al-Garni4,8, Muneera Altaweel3,4, Muhammad Aamir Khan11, Muhammad Farooq Sabar11 and Zafar Iqbal2,4,5,9,10,11*2Department of Medicine, College of Applied Medical Sciences, King Saud Bin Abdulaziz University for Health Sciences, Riyadh, Saudi Arabia
3Department of Medicine, King Abdulaziz Hospital, Al-Ahsa, Saudi Arabia
4King Abdullah International Medical Research Center (KAIMRC), Riyadh, Saudi Arabia
5Department of Biotechnology, Qarshi Univeristy, Lahore, Pakistan
6Department of Pediatrics, King Abdulaziz Hospital, College of Applied Medical Sciences, King Saud Bin Abdulaziz University for Health Sciences, Al-Ahsa, Saudi Arabia
7Department of Clinical Laboratory, Johns Hopkins Aramco HealthCare (JHAH), Alahsa, Saudi Arabia
8Department of Medicine, King Abdulaziz Hospital, College of Applied Medical Sciences, King Saud Bin Abdulaziz University for Health Sciences, Al-Ahsa, Saudi Arabia
9King Abdullah International Medical Research Centre (KAIMRC), College of Applied Medical Sciences, King Saud Bin Abdulaziz University for Health Sciences (KSAU-HS), Al-Ahsa, Saudi Arabia
10Pakistan Society for Molecular and Clinical Hematology, Lahore, Pakistan
11Hematology, Oncology & Pharmacogenetic Engineering Sciences Group (HOPES), Centre for Applied Molecular Biology (CAMB), University of the Punjab, Lahore, Pakistan
Received: 12-Aug-2024, Manuscript No. CPECR-24-26746; Editor assigned: 14-Aug-2024, Pre QC No. CPECR-24-26746 (PQ); Reviewed: 28-Aug-2024, QC No. CPECR-24-26746; Revised: 04-Sep-2024, Manuscript No. CPECR-24-26746 (R); Published: 11-Sep-2024, DOI: 10.35248/2161- 1459.24.14.431
Abstract
Inherited cardiomyopathies are a diverse group of heart muscle diseases caused by genetic mutations that result in structural and functional abnormalities of the myocardium. Understanding genotype-phenotype correlations in these conditions is vital for personalized cardiac medicine, enabling targeted therapeutic strategies and predictive diagnostics. This review explores the major types of inherited cardiomyopathies: Hypertrophic Cardiomyopathy (HCM), Dilated Cardiomyopathy (DCM), Arrhythmogenic Cardiomyopathy (ACM) and Restrictive Cardiomyopathy (RCM) and provides detailed insights into how different genetic mutations manifest as clinical features. The integration of multi-omics approaches and advanced disease modelling techniques has enhanced our ability to dissect these correlations. The review also discusses the implications of these findings for personalized medicine, including tailored therapeutic strategies, predictive diagnostics, and future research directions.
Keywords
Cardiomyopathy; Multi-omics techniques; Personalized medicine; Genetic testing
Introduction
Inherited cardiomyopathies represent a heterogeneous group of heart muscle diseases characterized by structural and functional abnormalities of the myocardium [1]. These conditions are primarily caused by genetic mutations that lead to varying clinical phenotypes. Understanding the genotype-phenotype correlations in these diseases is crucial for advancing personalized cardiac medicine, as it allows for the development of targeted therapeutic strategies and predictive diagnostics [2]. The integration of multi- omics approaches and advanced disease modelling techniques has further enhanced our ability to dissect these correlations and tailor therapeutic interventions [3]. Cardiomyopathies were first described in the mid-20th century, with early classifications based on clinical and pathological findings. Over time, it became evident that many cardiomyopathies had a genetic basis, leading to the identification of specific mutations associated with different forms of the disease. The major types of inherited cardiomyopathies include Hypertrophic Cardiomyopathy (HCM), Dilated Cardiomyopathy (DCM), Arrhythmogenic Cardiomyopathy (ACM), and Restrictive Cardiomyopathy (RCM) [4]. Genotype-phenotype correlations provide insights into how specific genetic mutations manifest as particular clinical features [2]. These correlations are essential for genetic counseling, risk stratification, and the development of personalized treatment plans.
Literature Review
Inherited cardiomyopathies
Inherited cardiomyopathies include Hypertrophic Cardiomyopathy (HCM) with thickened heart walls, Dilated Cardiomyopathy (DCM) with enlarged and weakened ventricles, Arrhythmogenic Cardiomyopathy (ACM) involving fibrofatty tissue in the heart, and Restrictive Cardiomyopathy (RCM) characterized by stiffened heart muscles, all leading to impaired cardiac function and increased risk of heart failure.
Hypertrophic Cardiomyopathy (HCM): HCM is the most common inherited cardiomyopathy, with a prevalence of 1 in 500 individuals. It is primarily caused by mutations in genes encoding sarcomeric proteins, such as MYH7, MYBPC3, and TNNT2. These mutations lead to abnormal sarcomeric function and myocardial hypertrophy [5]. Genotype-phenotype studies have shown that specific mutations are associated with varying degrees of hypertrophy, arrhythmias, and risk of sudden cardiac death [6]. For instance, mutations in the MYH7 gene often result in more severe hypertrophy and a higher risk of sudden cardiac death, necessitating aggressive management and consideration for Implantable Cardioverter-Defibrillators (ICDs) [7]. On the other hand, mutations in the MYBPC3 gene may present with milder hypertrophy but carry a higher risk of heart failure [8]. Recent clinical guidelines recommend genetic testing for all first-degree relatives of patients with HCM to identify those at risk and guide early intervention strategies [9]. Patients with MYH7 mutations often exhibit severe hypertrophy and are at a higher risk of sudden cardiac death [10], while MYBPC3 mutations are associated with a more diverse clinical presentation, ranging from mild to severe hypertrophy [8]. Clinical management may involve the use of ICDs for primary prevention in these patients. Additionally, beta-blockers and calcium channel blockers are recommended to manage symptoms and reduce arrhythmias [11].
Dilated Cardiomyopathy (DCM): DCM is characterized by left ventricular dilatation and systolic dysfunction [12]. It is caused by mutations in a diverse set of genes, including those encoding cytoskeletal, sarcomeric, and nuclear envelope proteins. Notable genes include TTN, LMNA, FLNC, DES, TMEM43 and DSP [13]. Genotype-phenotype correlations in DCM are complex, with some mutations leading to severe dilatation and heart failure, while others result in milder phenotypes [14,15]. TTN truncating variants are among the most common mutations found in DCM and are associated with a wide spectrum of clinical outcomes, from asymptomatic carriers to severe heart failure requiring transplantation [16,17].
Mutations in the LMNA gene are particularly associated with early-onset heart failure, conduction system disease, and arrhythmias, often necessitating the early implantation of pacemakers or ICDs [18,19]. Clinical guidelines emphasize the importance of early genetic testing in DCM patients to identify those with LMNA mutations and implement appropriate monitoring and preventive measures [20]. Therapies with DCM may vary by presciber, symptoms, and specific disease pathology. Angiotensin-converting enzyme inhibitors, Angiotensin II receptor blockers, β-blockers, mineralocorticoid receptor antagonists, ivabradine, and sodium- glucose contransporter-2 inhibitor showed a significant reduction of hospitalization and/or mortality. Other treatment options include pacemaker, ablation therapy, cardiac resynchronization therapy, and ICD therapy. Severe cases may require heart transplant, IV inotropes, durable ventricular assist device and extracorporeal membrane oxygenation [21].
Arrhythmogenic Cardiomyopathy (ACM): ACM or previously known as Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) is characterized by fibrofatty replacement of the right ventricular myocardium and ventricular arrhythmias. It is commonly caused by mutations in desmosomal genes, such as PKP2, DSP, and SG2. These mutations disrupt cell-cell adhesion, leading to myocardial degeneration and arrhythmias [22]. Genotype-phenotype studies have shown that specific mutations are associated with varying degrees of ventricular dysfunction and arrhythmic risk [23]. PKP2 mutations are often associated with more severe arrhythmic events and a higher risk of sudden cardiac death, necessitating aggressive arrhythmia management and consideration for ICD implantation [24]. DSP mutations may present with both right and left ventricular involvement, complicating the clinical picture and requiring comprehensive management strategies [25,26]. Current clinical guidelines recommend genetic testing for all first-degree relatives of ARVC patients to identify those at risk and guide surveillance and treatment plans [26,27]. Treating ARVC can be challenging due to progressive fibrosis, phenotypic variations, and differential diagnosis with other conditions. Patients with PKP2 mutations often exhibit severe ventricular arrhythmias, and clinical guidelines recommend the use of ICDs for primary prevention of sudden cardiac death. Antiarrhythmic drugs and beta-blockers are also recommended to manage arrhythmias. Patients with DSP mutations may require more comprehensive management due to potential involvement of both ventricles. Clinical management may involve the regular monitoring of both right and left ventricular function and the use of ICDs for arrhythmia prevention [22,28].
Restrictive Cardiomyopathy (RCM): RCM is the least common form of inherited cardiomyopathy, characterized by diastolic dysfunction and restrictive filling of the ventricles (24). It can be caused by mutations in sarcomeric and nonsarcomeric genes, including TNNI3, MYH7, FLNC, and DES [15,29].
Genotype-phenotype correlations in RCM are less well-defined due to the rarity of the disease, but certain mutations are associated with severe diastolic dysfunction and poor prognosis [30,31]. TNNI3 mutations can lead to severe restrictive physiology and early heart failure [32,33], while MYH7 mutations may present with overlapping features of HCM and RCM, complicating the diagnosis and management [34]. Clinical guidelines recommend comprehensive genetic testing in RCM patients to identify causative mutations and tailor treatment strategies accordingly [35]. Patients with TNNI3 mutations often present with severe diastolic dysfunction and may benefit from aggressive diuretic therapy to manage symptoms of heart failure [36]. Anticoagulation therapy may be recommended to prevent thromboembolic complications [37]. Due to the potential overlap with HCM [38], patients with MYH7 mutations may require a combination of therapies used for both HCM and RCM, including beta-blockers, calcium channel blockers, and diuretics. Regular monitoring for changes in phenotype and heart failure progression is crucial [39].
The layering of genotype and phenotype is complicated by overlaps amongst the major subtypes of inherited cardiomyopathies. Differential mutation detection in different types of cardiomyopathies can aid in differential diagnosis, prognostic stratification, and selection of treatment. For example, identifying a TTN mutation in a patient with DCM can indicate a potential for severe heart failure and guide decisions regarding early intervention with heart failure therapies or consideration for heart transplantation [40]. Similarly, detecting a PKP2 mutation in a patient with ARVC can highlight the need for aggressive arrhythmia management and consideration of an ICD [41].
Multi-omics approaches in cardiomyopathies
The advent of multi-omics technologies has revolutionized the study of cardiomyopathies [42]. Multi-omics approaches integrate data from various omics layers, including genomics, transcriptomics, proteomics, and metabolomics, to provide a comprehensive view of the molecular mechanisms driving disease phenotypes [43].
Genomics: Genomic studies have identified numerous genetic mutations associated with inherited cardiomyopathies. Whole- Exome Sequencing (WES) and Whole-Genome Sequencing (WGS) have been particularly useful in identifying rare and novel variants [44]. These technologies have also facilitated the study of genetic modifiers that influence disease severity and penetrance [45].
Transcriptomics: Transcriptomic analyses, such as RNA sequencing, provide insights into gene expression changes associated with cardiomyopathies [46,47]. These studies have identified differentially expressed genes and pathways that contribute to disease pathogenesis [48].
For example, transcriptomic studies in HCM have revealed upregulation of hypertrophic signaling pathways and downregulation of energy metabolism genes [49].
Proteomics: Proteomic approaches, including mass spectrometry, have been used to study protein expression and post-translational modifications in cardiomyopathies [50,51]. These studies have identified altered protein networks and signaling pathways that contribute to disease phenotypes [52]. For instance, proteomic analyses in DCM have revealed dysregulation of cytoskeletal and mitochondrial proteins [53].
Metabolomics: Metabolomic studies provide insights into metabolic alterations associated with cardiomyopathies [50,54]. These studies have identified changes in metabolites and metabolic pathways that contribute to disease progression [55]. For example, metabolomics analyses in ARVC have revealed alterations in lipid metabolism and energy production [56].
Disease modeling in cardiomyopathies
Advanced disease modeling techniques, such as the use of induced Pluripotent Stem Cells (iPSCs) and animal models, have provided valuable platforms for studying cardiomyopathies. These models enable the investigation of disease mechanisms, the testing of therapeutic interventions, and the study of patient-specific disease phenotypes [57]. Indeed, these preclinical models can also help to clarify the genotype-phenotype associations and the pathophysiological mechanisms.
Induced Pluripotent Stem Cells (iPSCs): iPSCs are derived from patient-specific somatic cells and can be differentiated into cardiomyocytes [58]. While monolayer iPSC cultures exhibit considerable scalability, they yield a model that is inherently simplified and less intricate in comparison to the complexity observed in both 3D in vitro models and in vivo systems (Figure 1). Despite this, iPSC-derived cardiomyocytes can still recapitulate many aspects of the patient’s disease phenotype, allowing for the study of disease mechanisms and drug testing in a patient-specific context. For example, iPSC models of HCM have been used to study the effects of MYBPC3 mutations on sarcomere function and calcium handling [59]. Indeed, the technology of using iPSC-derived cardiomyocytes has been used to model numerous inherited cardiomyopathies, whether by producing patient-specific cells, editing the genome of healthy cells, or even overexpressing mutated ion channels.
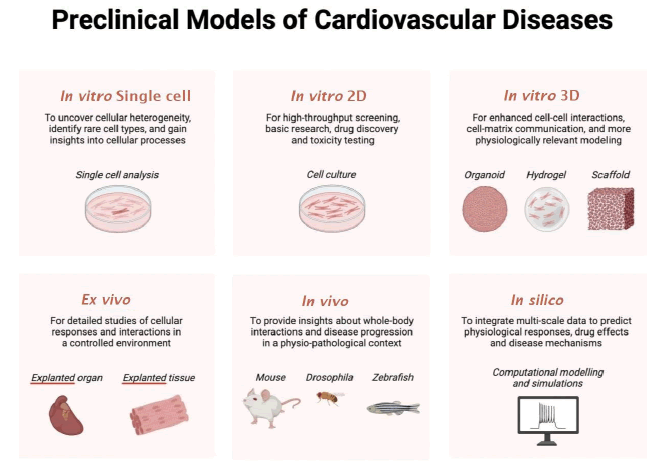
Figure 1: Summary of the available in vitro, in vivo, ex vivo, and in silico platforms that can model inherited cardiomyopathy.
Animal models: Animal models, including transgenic mice and zebrafish, have been widely used to study cardiomyopathies [60]. These models allow for the investigation of gene function and the study of disease progression in a whole-organism context [61]. For instance, transgenic mouse models of DCM have been used to study the effects of TTN mutations on cardiac function and remodeling [62].
Discussion
Future impact of cardiac regenerative medicine in finding innovative treatment for cardiomyopathies
The field of cardiac regenerative medicine and the use of cardiac stem cells hold significant promise for developing innovative treatments for cardiomyopathies. Recent advancements in stem cell biology and regenerative medicine have paved the way for novel therapeutic approaches that aim to repair or replace damaged cardiac tissue, thereby improving cardiac function and patient outcomes [63].
Cardiac stem cells, including induced Pluripotent Stem Cells (iPSCs) and Mesenchymal Stem Cells (MSCs), have shown potential in regenerating damaged myocardium [64]. iPSCs can be generated from a patient’s own somatic cells and differentiated into cardiomyocytes, which can then be used to replace damaged or dysfunctional heart tissue. This approach not only provides a personalized therapy option but also minimizes the risk of immune rejection [65,66]. Several studies have demonstrated the efficacy of iPSC-derived cardiomyocytes in improving cardiac function in animal models of cardiomyopathy [67].
In addition to iPSCs, MSCs have been explored for their regenerative potential in cardiac tissues. MSCs possess immunomodulatory properties and can promote tissue repair through paracrine signaling. They secrete a variety of growth factors, cytokines, and extracellular vesicles that can enhance angiogenesis, reduce fibrosis, and stimulate endogenous cardiac repair mechanisms [68]. Clinical trials investigating the use of MSCs in patients with cardiomyopathy have shown promising results, with improvements in cardiac function and reductions in adverse remodeling [69,70].
Another promising approach in cardiac regenerative medicine is the use of Extracellular Vesicles (EVs) derived from stem cells. EVs are small membrane-bound vesicles that contain bioactive molecules, such as proteins, lipids, and nucleic acids, which can modulate cellular processes and promote tissue repair. Studies have shown that EVs derived from stem cells can enhance cardiac regeneration by promoting cell survival, reducing inflammation, and stimulating angiogenesis [71]. The use of EVs as a therapeutic modality offers several advantages, including ease of administration, low immunogenicity, and the ability to cross biological barriers [72,73].
Tissue engineering and 3D bioprinting technologies are also being explored for their potential to create functional cardiac tissues for transplantation. These technologies involve the use of biomaterials, stem cells, and growth factors to construct three-dimensional cardiac tissues that can mimic the structure and function of native myocardium. Engineered cardiac tissues can be used to replace damaged heart tissue and restore cardiac function [74]. Recent advancements in 3D bioprinting have enabled the fabrication of complex cardiac structures with high precision, which can be tailored to individual patients’ needs [75,76].
Despite the promising potential of cardiac regenerative medicine, several challenges remain to be addressed. These include the scalability and reproducibility of stem cell production, the risk of tumorigenicity, and the integration of transplanted cells with the host myocardium [77]. Additionally, the long-term safety and efficacy of these therapies need to be thoroughly evaluated in clinical trials [78].
Future research in cardiac regenerative medicine should focus on optimizing cell delivery methods, enhancing the survival and integration of transplanted cells, and developing strategies to modulate the cardiac microenvironment to support regeneration [79,80]. Collaborative efforts between researchers, clinicians, and industry partners will be essential to translate these innovative therapies from the bench to the bedside.
Implications for personalized medicine
Personalized medicine aims to tailor medical treatment to the individual characteristics of each patient [81]. In the context of cardiomyopathies, understanding genotype-phenotype correlations allows for the development of targeted therapies and predictive diagnostics [35].
Tailored therapeutic strategies: Personalized treatment plans based on genetic profiles have shown promise in improving outcomes for patients with cardiomyopathies [82,83]. For example, genotype-specific therapies, such as small-molecule inhibitors and gene therapies, are being developed for patients with specific mutations [84]. Additionally, personalized exercise and lifestyle recommendations can be made based on the patient’s genetic risk factors [85].
Predictive diagnostics: Predictive diagnostics based on genotype- phenotype correlations can help identify individuals at high risk for developing cardiomyopathies and guide early intervention strategies [86,87]. For example, genetic testing can identify individuals with pathogenic mutations, allowing for close monitoring and preventive measures to reduce the risk of adverse outcomes [88].
Challenges and future directions: Despite the promise of personalized medicine, there are several challenges to its implementation. These include the complexity of genotype-phenotype correlations, the need for large-scale data integration, and the ethical considerations of genetic testing [81,89,90]. Future research should focus on addressing these challenges and further refining personalized treatment and diagnostic approaches.
Gaps in current research
Despite significant advancements, there are still gaps in our understanding of genotype-phenotype correlations in cardiomyopathies. Some genetic mutations have variable expressivity and incomplete penetrance, complicating the prediction of clinical outcomes [91]. Additionally, the integration of multi-omics data remains challenging due to the complexity and volume of data generated [92]. The lack of knowledge can be summarized into:
Unresolved correlations: Certain genotype-phenotype correlations remain unclear, with some mutations leading to highly variable clinical presentations. For example, mutations in the LMNA gene can cause a wide range of phenotypes, from mild dilatation to severe heart failure and arrhythmias [93]. Further research is needed to elucidate the factors that contribute to this variability.
Technological and methodological limitations: Current research methodologies have limitations that hinder the full understanding of genotype-phenotype correlations. For instance, WES and WGS may miss certain types of genetic variants, such as structural variants and deep intronic mutations [94]. Additionally, the integration of multi-omics data requires advanced computational tools and techniques, which are still in development [95].
Recommendations for future research: To address these gaps, future research should focus on improving genetic testing techniques, developing better data integration methods, and conducting large-scale studies to validate genotype-phenotype correlations. Collaborative efforts and the use of advanced technologies, such as machine learning, will be essential in advancing our understanding of inherited cardiomyopathies.
Future directions
Advances in multi-omics: Emerging technologies in multi- omics research, such as single-cell RNA sequencing and spatial transcriptomics, have the potential to provide deeper insights into the cellular and molecular mechanisms of cardiomyopathies [96]. These technologies can help identify novel therapeutic targets and biomarkers for personalized treatment [97].
Innovations in disease modelling: New approaches to disease modeling, such as the use of organ-on-a-chip technology and 3D bioprinting, can enhance our ability to study cardiomyopathies in a more physiologically relevant context [98]. These models can be used to test the efficacy and safety of new therapies and to study the interactions between different cell types in the heart [99].
Translational research and clinical applications: Bridging the gap between research and clinical practice is essential for translating scientific discoveries into effective treatments for patients with cardiomyopathies [100]. Collaborative efforts between researchers, clinicians, and industry partners will be important in developing and implementing personalized therapeutic strategies.
Conclusion
In summary, genotype-phenotype correlations play a pivotal role in the study and management of inherited cardiomyopathies. The integration of multi-omics approaches and advanced disease modeling techniques has significantly advanced our understanding of these complex diseases. However, further research is needed to fully elucidate the molecular mechanisms driving disease phenotypes and to translate these findings into effective personalized therapeutic strategies. Continued advancements in genetic testing, data integration, and disease modeling will be essential in improving outcomes for patients with inherited cardiomyopathies. Additionally, the emerging therapeutic strategies in cardiac regenerative medicine and the role of gene editing technologies such as CRISPR-Cas9 offer promising avenues for treatment. Advances in cardiac imaging, artificial intelligence, and the use of stem cell-derived extracellular vesicles may further enhance our ability to diagnose and treat these conditions. The integration of personalized medicine, novel biomarkers, and wearable technology into clinical practice will play an important role in managing cardiomyopathies. Future research should continue to focus on these innovative approaches to develop more effective treatments and improve patient outcomes.
Acknowledgments
The graphical abstract was created with BioRender.com. We acknowledge the Institutional Review Board (IRB) of the King Abdullah International Medical Research Center (KAIMRC), National Guard Health Affairs, Saudi Arabia, for ethical approval of this project (RYD-24-417780-88619) dated 02 June 2024, although this research received no funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lukas Laws J, Lancaster MC, Ben Shoemaker M, Stevenson WG, Hung RR, Wells Q, et al. Arrhythmias as presentation of genetic cardiomyopathy. Circ Res. 2022;130(11):1698-1722.
[Crossref] [Google Scholar] [PubMed]
- Fan X, Yang G, Duru F, Grilli M, Akin I, Zhou X, et al. Arrhythmogenic cardiomyopathy: From preclinical models to genotype-phenotype correlation and pathophysiology. Stem Cell Rev Rep. 2023;19(8):2683-2708.
[Crossref] [Google Scholar] [PubMed]
- Wu Y, Xie L. AI-driven multi-omics integration for multi-scale predictive modeling of causal genotype-environment-phenotype relationships. arXiv preprint. 2024.
- Bueno CO, Jurado ML. Cardiomyopathies: A historical journey. 2024.
- Members WC, Ommen SR, Ho CY, Asif IM, Balaji S, Burke MA, et al. 2024 AHA/ACC/AMSSM/HRS/PACES/SCMR guideline for the management of hypertrophic cardiomyopathy: A report of the American heart association/American college of cardiology joint committee on clinical practice guidelines. J Am Coll Cardiol. 2024;83(23):2324-2405.
[Crossref] [Google Scholar] [PubMed]
- Lopes LR, Ho CY, Elliott PM. Genetics of hypertrophic cardiomyopathy: Established and emerging implications for clinical practice. Eur Heart J. 2024;45(30):2727-2734.
[Crossref] [Google Scholar] [PubMed]
- Franke M, Książczyk TM, Dux M, Chmielewski P, Truszkowska G, Czapczak D, et al. A MYH7 variant in a five-generation-family with hypertrophic cardiomyopathy. Front Genet. 2024;15:1306333.
[Crossref] [Google Scholar] [PubMed]
- Ananthamohan K, Stelzer JE, Sadayappan S. Hypertrophic cardiomyopathy in MYBPC3 carriers in aging. J Cardiovasc Aging. 2024;4(1):9.
[Crossref] [Google Scholar] [PubMed]
- Ireland CG, Ho CY. Genetic testing in hypertrophic cardiomyopathy. Am J Cardiol. 2024;212:S4-S13.
[Crossref] [Google Scholar] [PubMed]
- Jansen M, de Brouwer R, Hassanzada F, Schoemaker AE, Schmidt AF, Kooijman-Reumerman MD, et al. Penetrance and prognosis of MYH7 variant-associated cardiomyopathies: Results from a Dutch multicenter cohort study. JACC Heart Fail. 2024;12(1):134-147.
[Crossref] [Google Scholar] [PubMed]
- Hutt E, Desai MY. Medical treatment strategies for hypertrophic cardiomyopathy. Am J Cardiol. 2024;212:S33-S41.
[Crossref] [Google Scholar] [PubMed]
- Eda Y, Nabeta T, Iikura S, Takigami Y, Fujita T, Iida Y, et al. Non‐dilated left ventricular cardiomyopathy vs. dilated cardiomyopathy: Clinical background and outcomes. ESC Heart Fail. 2024.
[Crossref] [Google Scholar] [PubMed]
- Kärkkäinen S, Peuhkurinen K. Genetics of dilated cardiomyopathy. Ann Med. 2007;39(2):91-107.
[Crossref] [Google Scholar] [PubMed]
- Voinescu OR, Ionescu BI, Militaru S, Afana AS, Sascau R, Vasiliu L, et al. Genetic characterization of dilated cardiomyopathy in romanian adult patients. Int J Mol Sci. 2024;25(5):2562.
[Crossref] [Google Scholar] [PubMed]
- Voinescu OR, Ionac A, Sosdean R, Ionac I, Ana LS, Kundnani NR, et al. Genotype-phenotype insights of inherited cardiomyopathies-A review. Medicina. 2024;60(4):543.
[Crossref] [Google Scholar] [PubMed]
- Irene B, Carla G, Pia CM, Gemma PM, Fiammetta A, Gabriella P, et al. Dilated cardiomyopathy due to a novel combination of TTN and BAG3 genetic variants: From acute heart failure to subclinical phenotypes. Cardiovasc Pathol. 2024:107675.
[Crossref] [Google Scholar] [PubMed]
- León P, Franco P, Hinojosa N, Torres K, Moreano A, Romero VI. TTN novel splice variant in familial dilated cardiomyopathy and splice variants review: A case report. Front Cardiovasc Med. 2024;11:1387063.
[Crossref] [Google Scholar] [PubMed]
- Mariani MV, Pierucci N, Fanisio F, Laviola D, Silvetti G, Piro A, et al. Inherited arrhythmias in the pediatric population: An updated overview. Medicina (Kaunas). 2024;60(1):94.
[Crossref] [Google Scholar] [PubMed]
- Blich M, Darawsha W, Eyal A, Shehadeh F, Boulous M, Gepstein L, et al. The role of early cardiac resynchronization therapy implantation in dilated cardiomyopathy patients with narrow QRS carrying lamin A/C mutation. Am J Cardiovasc Dis. 2024;14(1):47.
[Crossref] [Google Scholar] [PubMed]
- Arnautu DA, Cozma D, Lala IR, Arnautu SF, Tomescu MC, Andor M. Risk Assessment and Personalized Treatment Options in Inherited Dilated Cardiomyopathies: A Narrative Review. Biomedicines. 2024;12(8):1643.
- Malinow I, Fong DC, Miyamoto M, Badran S, Hong CC. Pediatric dilated cardiomyopathy: A review of current clinical approaches and pathogenesis. Front Pediatr. 2024;12:1404942.
[Crossref] [Google Scholar] [PubMed]
- Al-Aidarous S, Protonotarios A, Elliott PM, Lambiase PD. Management of arrhythmogenic right ventricular cardiomyopathy. Heart. 2024;110(3):156-162.
[Crossref] [Google Scholar] [PubMed]
- La Vecchia G, Bisaccia G, Perotto M, Leo I, Procopio MC, Licordari R, et al. Redefining genotype-phenotype correlation among patients with arrhythmogenic cardiomyopathy: A cardiovascular magnetic resonance cohort study. J Cardiovasc Magn Reson. 2024;26.
- Phan DP, Tran TV, Le VK, Nguyen TV. Detection of gene mutation in the prognosis of a patient with arrhythmogenic right ventricular cardiomyopathy: A case report. J Med Case Rep. 2024;18(1):49.
[Crossref] [Google Scholar] [PubMed]
- Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation. 2020;141(23):1872-1884.
[Crossref] [Google Scholar] [PubMed]
- Cianci V, Forzese E, Sapienza D, Cianci A, Ieni A, Germanà A, et al. Arrhythmogenic right ventricular cardiomyopathy post-mortem assessment: A systematic review. Int J Mol Sci. 2024;25(5):2467.
[Crossref] [Google Scholar] [PubMed]
- Hespe S, Gray B, Puranik R, Peters S, Sweeting J, Ingles J. The role of genetic testing in management and prognosis of individuals with inherited cardiomyopathies. Trends Cardiovasc Med. 2024;12:S1050.
[Crossref] [Google Scholar] [PubMed]
- Moisa SM, Spoiala EL, Cinteza E, Vatasescu R, Butnariu LI, Brinza C, et al. Arrhythmogenic right ventricular cardiomyopathy in children: A systematic review. Diagnostics (Basel). 2024;14(2):175.
[Crossref] [Google Scholar] [PubMed]
- Cimiotti D, Budde H, Hassoun R, Jaquet K. Genetic restrictive cardiomyopathy: Causes and consequences-an integrative approach. Int J Mol Sci. 2021;22(2):558.
[Crossref] [Google Scholar] [PubMed]
- Rapezzi C, Aimo A, Barison A, Emdin M, Porcari A, Linhart A, et al. Restrictive cardiomyopathy: Definition and diagnosis. Eur Heart J. 2022;43(45):4679-4693.
[Crossref] [Google Scholar] [PubMed]
- Yang Z, Chen J, Li H, Lin Y. Genotype-phenotype associations with restrictive cardiomyopathy induced by pathogenic genetic mutations. Rev Cardiovasc Med. 2022;23(6):185.
[Crossref] [Google Scholar] [PubMed]
- Sorrentino U, Gabbiato I, Canciani C, Calosci D, Rigon C, Zuccarello D, et al. Homozygous TNNI3 mutations and severe early onset dilated cardiomyopathy: Patient report and review of the literature. Genes (Basel). 2023;14(3):748.
[Crossref] [Google Scholar] [PubMed]
- Jin Y, Xu J, Hua Y, Zhang H, Li Y. Challenging of ECMO application in pediatric restrictive cardiomyopathy: Case report of a novel TNNI3 variant. Front Cardiovasc Med. 2024;11:1365209.
[Crossref] [Google Scholar] [PubMed]
- Catrina BI, Batar F, Baltat G, Bitea CI, Puia A, Stoia O, et al. A family with MYH7 mutation and different forms of cardiomyopathies. Biomedicines. 2023;11(7):2065.
[Crossref] [Google Scholar] [PubMed]
- Yogasundaram H, Alhumaid W, Dzwiniel T, Christian S, Oudit GY. Cardiomyopathies and genetic testing in heart failure: Role in defining phenotype-targeted approaches and management. Can J Cardiol. 2021;37(4):547-559.
[Crossref] [Google Scholar] [PubMed]
- Ueno M, Takeda A, Yamazawa H, Takei K, Furukawa T, Suzuki Y, et al. A case report: Twin sisters with restrictive cardiomyopathy associated with rare mutations in the cardiac troponin I gene. J Cardiol Cases. 2021;23(4):154-157.
[Crossref] [Google Scholar] [PubMed]
- Dai HL, Wang QH, Su X, Ding YC, Guang XF. Pediatric restrictive cardiomyopathy: A case report. J Int Med Res. 2023;51(8):03000605231188276.
[Crossref] [Google Scholar] [PubMed]
- Vepsäläinen T, Heliö T, Vasilescu C, Martelius L, Weckström S, Koskenvuo J, et al. MYH7 genotype-phenotype correlation in a cohort of Finnish patients. Cardiogenetics. 2022;12(1):122-132.
- Kian W, Zemel M, Kestenbaum EH, Alguayn W, Shvarts B, Sharb AA, et al. Cardiomyopathy etiologies, symptoms and management. In Cardiomyopathy-Disease of the Heart Muscle. IntechOpen. 2021.
- Akhtar MM, Lorenzini M, Cicerchia M, Ochoa JP, Hey TM, Sabater Molina M, et al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circ Heart Fail. 2020;13(10):e006832.
[Crossref] [Google Scholar] [PubMed]
- Spadotto A, Morabito D, Carecci A, Massaro G, Statuto G, Angeletti A, et al. The challenges of diagnosis and treatment of arrhythmogenic cardiomyopathy: Are we there yet?. Rev Cardiovasc Med. 2022;23(8):283.
[Crossref] [Google Scholar] [PubMed]
- Wang RS, Maron BA, Loscalzo J. Multiomics network medicine approaches to precision medicine and therapeutics in cardiovascular diseases. Arterioscler Thromb Vasc Biol. 2023;43(4):493-503.
[Crossref] [Google Scholar] [PubMed]
- Doran S, Arif M, Lam S, Bayraktar A, Turkez H, Uhlen M, et al. Multi-omics approaches for revealing the complexity of cardiovascular disease. Brief Bioinform. 2021;22(5):bbab061.
[Crossref] [Google Scholar] [PubMed]
- Kim MJ, Yum MS, Seo GH, Lee Y, Jang HN, Ko TS, et al. Clinical application of whole exome sequencing to identify rare but remediable neurologic disorders. J Clin Med. 2020;9(11):3724.
[Crossref] [Google Scholar] [PubMed]
- Brlek P, Bulic L, Bračić M, Projić P, Škaro V, Shah N, et al. Implementing Whole Genome Sequencing (WGS) in clinical practice: Advantages, challenges, and future perspectives. Cells. 2024;13(6):504.
[Crossref] [Google Scholar] [PubMed]
- Wehrens M, de Leeuw AE, Wright-Clark M, Eding JE, Boogerd CJ, Molenaar B, et al. Single-cell transcriptomics provides insights into hypertrophic cardiomyopathy. Cell Rep. 2022;39(6).
[Crossref] [Google Scholar] [PubMed]
- Simonson B, Chaffin M, Hill MC, Atwa O, Guedira Y, Bhasin H, et al. Single-nucleus RNA sequencing in ischemic cardiomyopathy reveals common transcriptional profile underlying end-stage heart failure. Cell Rep. 2023;42(2):112086.
[Crossref] [Google Scholar] [PubMed]
- Li CX, Chen J, Lv SK, Li JH, Li LL, Hu X. Whole‐transcriptome RNA sequencing reveals significant differentially expressed mRNAs, miRNAs, and lncRNAs and related regulating biological pathways in the peripheral blood of COVID‐19 patients. Mediators Inflamm. 2021;2021(1):6635925.
[Crossref] [Google Scholar] [PubMed]
- Chen S, Hu J, Xu Y, Yan J, Li S, Chen L, et al. Transcriptome analysis of human hypertrophic cardiomyopathy reveals inhibited cardiac development pathways in children. iScience. 2024;27(1):108642.
[Crossref] [Google Scholar] [PubMed]
- Previs MJ, O’Leary TS, Morley MP, Palmer BM, LeWinter M, Yob JM, et al. Defects in the proteome and metabolome in human hypertrophic cardiomyopathy. Circ Heart Fail. 2022;15(6):e009521.
[Crossref] [Google Scholar] [PubMed]
- Yuan L, Bu S, Du M, Wang Y, Ju C, Huang D, et al. RNF207 exacerbates pathological cardiac hypertrophy via post-translational modification of TAB1. Cardiovasc Res. 2023;119(1):183-194.
[Crossref] [Google Scholar] [PubMed]
- Shimada YJ, Raita Y, Liang LW, Maurer MS, Hasegawa K, Fifer MA, et al. Comprehensive proteomics profiling reveals circulating biomarkers of hypertrophic cardiomyopathy. Circ Heart Fail. 2021;14(7):e007849.
[Crossref] [Google Scholar] [PubMed]
- Liu M, Zhai L, Yang Z, Li S, Liu T, Chen A, et al. Integrative proteomic analysis reveals the cytoskeleton regulation and mitophagy difference between ischemic cardiomyopathy and dilated cardiomyopathy. Mol Cell Proteomics. 2023;22(12):100667.
[Crossref] [Google Scholar] [PubMed]
- Gonzalez-Covarrubias V, Martínez-Martínez E, Del Bosque-Plata L. The potential of metabolomics in biomedical applications. Metabolites. 2022;12(2):194.
[Crossref] [Google Scholar] [PubMed]
- Lopaschuk GD, Karwi QG, Tian R, Wende AR, Abel ED. Cardiac energy metabolism in heart failure. Circ Res. 2021;128(10):1487-1513.
[Crossref] [Google Scholar] [PubMed]
- Song R, Wu H, Yu L, Yu J, Yang W, Wu W, et al. Multiomics analysis reveals extensive remodeling of the extracellular matrix and cellular metabolism due to plakophilin-2 knockdown in guinea pigs. bioRxiv. 2024.
- Kawaguchi N, Nakanishi T. Animal disease models and patient-iPS-cell-derived in vitro disease models for cardiovascular biology-how close to disease?. Biology (Basel). 2023;12(3):468.
[Crossref] [Google Scholar] [PubMed]
- Park S, Gwon Y, Khan SA, Jang KJ, Kim J. Engineering considerations of iPSC-based personalized medicine. Biomater Res. 2023;27(1):67.
[Crossref] [Google Scholar] [PubMed]
- Dababneh S, Hamledari H, Maaref Y, Jayousi F, Baygi DH, Khan A, et al. Advances in hypertrophic cardiomyopathy disease modeling using human iPSC-derived cardiomyocytes. Can J Cardiol. 2024;40(5):766-776.
[Crossref] [Google Scholar] [PubMed]
- Blackwell DJ, Schmeckpeper J, Knollmann BC. Animal models to study cardiac arrhythmias. Circ Res. 2022;130(12):1926-1964.
[Crossref] [Google Scholar] [PubMed]
- Niu Y, Sun Y, Liu Y, Du K, Xu X, Ding Y. Using zebrafish animal model to study the genetic underpinning and mechanism of arrhythmogenic cardiomyopathy. Int J Mol Sci. 2023;24(4):4106.
[Crossref] [Google Scholar] [PubMed]
- Inoue S, Ko T, Shindo A, Nomura S, Yamada T, Jimba T, et al. Association between clonal hematopoiesis and left ventricular reverse remodeling in nonischemic dilated cardiomyopathy. JACC Basic Transl Sci. 2024.
- Kasai-Brunswick TH, Carvalho AB, de Carvalho AC. Stem cell therapies in cardiac diseases: Current status and future possibilities. World J Stem Cells. 2021;13(9):1231-1247.
[Crossref] [Google Scholar] [PubMed]
- Neef K, Drey F, Lepperhof V, Wahlers T, Hescheler J, Choi YH, et al. Co-transplantation of mesenchymal stromal cells and induced pluripotent stem cell-derived cardiomyocytes improves cardiac function after myocardial damage. Front Cardiovasc Med. 2022;8:794690.
[Crossref] [Google Scholar] [PubMed]
- Fujita J, Tohyama S, Kanazawa H, Kishino Y, Okada M, Tanosaki S, et al. Clinical application of iPSC-derived cardiomyocytes in patients with advanced heart failure. In Advanced Technologies in Cardiovascular Bioengineering. Springer. 2022:361-374.
- Yoshida S, Miyagawa S, Toyofuku T, Fukushima S, Kawamura T, Kawamura A, et al. Syngeneic mesenchymal stem cells reduce immune rejection after induced pluripotent stem cell-derived allogeneic cardiomyocyte transplantation. Sci Rep. 2020;10(1):4593.
[Crossref] [Google Scholar] [PubMed]
- Vo QD, Saito Y, Nakamura K, Iida T, Yuasa S. Induced pluripotent stem cell-derived cardiomyocytes therapy for ischemic heart disease in animal model: A meta-analysis. Int J Mol Sci. 2024;25(2):987.
[Crossref] [Google Scholar] [PubMed]
- Müller L, Tunger A, Wobus M, von Bonin M, Towers R, Bornhäuser M, et al. Immunomodulatory properties of mesenchymal stromal cells: An update. Front Cell Dev Biol. 2021;9:637725.
[Crossref] [Google Scholar] [PubMed]
- Yuce K. The application of mesenchymal stem cells in different cardiovascular disorders: Ways of administration, and the effectors. Stem Cell Rev Rep. 2024:1-21.
[Crossref] [Google Scholar] [PubMed]
- da Silva JS, Gonçalves RG, Vasques JF, Rocha BS, Nascimento-Carlos B, Montagnoli TL, et al. Mesenchymal stem cell therapy in diabetic cardiomyopathy. Cells. 2022;11(2):240.
[Crossref] [Google Scholar] [PubMed]
- Kou M, Huang L, Yang J, Chiang Z, Chen S, Liu J, et al. Mesenchymal stem cell-derived extracellular vesicles for immunomodulation and regeneration: A next generation therapeutic tool?. Cell Death Dis. 2022;13(7):580.
[Crossref] [Google Scholar] [PubMed]
- de Jong B, Barros ER, Hoenderop JG, Rigalli JP. Recent advances in extracellular vesicles as drug delivery systems and their potential in precision medicine. Pharmaceutics. 2020;12(11):1006.
[Crossref] [Google Scholar] [PubMed]
- Simeone P, Bologna G, Lanuti P, Pierdomenico L, Guagnano MT, Pieragostino D, et al. Extracellular vesicles as signaling mediators and disease biomarkers across biological barriers. Int J Mol Sci. 2020;21(7):2514.
[Crossref] [Google Scholar] [PubMed]
- Roacho-Pérez JA, Garza-Treviño EN, Moncada-Saucedo NK, Carriquiry-Chequer PA, Valencia-Gómez LE, Matthews ER, et al. Artificial scaffolds in cardiac tissue engineering. Life (Basel). 2022;12(8):1117.
[Crossref] [Google Scholar] [PubMed]
- Jafari A, Ajji Z, Mousavi A, Naghieh S, Bencherif SA, Savoji H. Latest advances in 3D bioprinting of cardiac tissues. Adv Mater Technol. 2022;7(11):2101636.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Wang L, Li T, Liu S, Guo B, Huang W, et al. 3D bioprinting in cardiac tissue engineering. Theranostics. 2021;11(16):7948.
[Crossref] [Google Scholar] [PubMed]
- Silver SE, Barrs RW, Mei Y. Transplantation of human pluripotent stem cell-derived cardiomyocytes for cardiac regenerative therapy. Front Cardiovasc Med. 2021;8:707890.
[Crossref] [Google Scholar] [PubMed]
- Margiana R, Markov A, Zekiy AO, Hamza MU, Al-Dabbagh KA, Al-Zubaidi SH, et al. Clinical application of mesenchymal stem cell in regenerative medicine: A narrative review. Stem Cell Res Ther. 2022;13(1):366.
[Crossref] [Google Scholar] [PubMed]
- Häneke T, Sahara M. Progress in bioengineering strategies for heart regenerative medicine. Int J Mol Sci. 2022;23(7):3482.
[Crossref] [Google Scholar] [PubMed]
- Mazzola M, Di Pasquale E. Toward cardiac regeneration: Combination of pluripotent stem cell-based therapies and bioengineering strategies. Front Bioeng Biotechnol. 2020;8:455.
[Crossref] [Google Scholar] [PubMed]
- Hassan M, Awan FM, Naz A, deAndrés-Galiana EJ, Alvarez O, Cernea A, et al. Innovations in genomics and big data analytics for personalized medicine and health care: A review. Int J Mol Sci. 2022;23(9):4645.
[Crossref] [Google Scholar] [PubMed]
- Crotti L, Brugada P, Calkins H, Chevalier P, Conte G, Finocchiaro G, et al. From gene-discovery to gene-tailored clinical management: 25 years of research in channelopathies and cardiomyopathies. Europace. 2023;25(8):euad180.
[Crossref] [Google Scholar] [PubMed]
- Elliott PM. Personalized medicine for dilated cardiomyopathy. Eur Heart J. 2021;42(2):175-177.
[Crossref] [Google Scholar] [PubMed]
- Chaput D, Andelfinger G. MEK inhibition for RASopathy-associated hypertrophic cardiomyopathy: Clinical application of a basic concept. Can J Cardiol. 2024;40(5):789-799.
[Crossref] [Google Scholar] [PubMed]
- Farrokhi M, Taheri F, Khouzani PJ, Rahmani E, Tavakoli R, Fard AM, et al. Role of precision medicine and personalized medicine in the treatment of diseases. Kindle. 2023;3(1):1-64.
- Paldino A, Dal Ferro M, Stolfo D, Gandin I, Medo K, Graw S, et al. Prognostic prediction of genotype vs. phenotype in genetic cardiomyopathies. J Am Coll Cardiol. 2022;80(21):1981-1994.
[Crossref] [Google Scholar] [PubMed]
- Njoroge JN, Mangena JC, Aribeana C, Parikh VN. Emerging Genotype-Phenotype associations in dilated cardiomyopathy. Curr Cardiol Rep. 2022;24(9):1077-1084.
[Crossref] [Google Scholar] [PubMed]
- Ahluwalia M, Ho CY. Cardiovascular genetics: The role of genetic testing in diagnosis and management of patients with hypertrophic cardiomyopathy. Heart. 2021;107(3):183-189.
[Crossref] [Google Scholar] [PubMed]
- Magrinelli F, Balint B, Bhatia KP. Challenges in clinicogenetic correlations: One gene-many phenotypes. Mov Disord Clin Pract. 2021;8(3):299-310.
[Crossref] [Google Scholar] [PubMed]
- Balogun OD, Ayo-Farai O, Ogundairo O, Maduka CP, Okongwu CC, Babarinde AO, et al. The role of pharmacists in personalised medicine: A review of integrating pharmacogenomics into clinical practice. Int Med Sci Res J. 2024;4(1):19-36.
- Kingdom R, Wright CF. Incomplete penetrance and variable expressivity: From clinical studies to population cohorts. Front Genet. 2022;13:920390.
[Crossref] [Google Scholar] [PubMed]
- Kuang Z, Kong M, Yan N, Ma X, Wu M, Li J. Precision cardio-oncology: Update on omics-based diagnostic methods. Curr Treat Options Oncol. 2024:1-23.
[Crossref] [Google Scholar] [PubMed]
- Tiwari V, Alam MJ, Bhatia M, Navya M, Banerjee SK. The structure and function of lamin A/C: Special focus on cardiomyopathy and therapeutic interventions. Life Sci. 2024:122489.
[Crossref] [Google Scholar] [PubMed]
- Maggi J, Koller S, Feil S, Bachmann-Gagescu R, Gerth-Kahlert C, Berger W. Limited added diagnostic value of whole genome sequencing in genetic testing of inherited retinal diseases in a Swiss patient cohort. Int J Mol Sci. 2024;25(12):6540.
[Crossref] [Google Scholar] [PubMed]
- Kaithal P, Kanchan S, Kesheri M. Recent advances in biological omics databases and tools in human health. In Microbial Omics in Environment and Health. 2024:311-341.
- Palmer JA, Rosenthal N, Teichmann SA, Litvinukova M. Revisiting cardiac biology in the era of single cell and spatial omics. Circ Res. 2024;134(12):1681-1702.
[Crossref] [Google Scholar] [PubMed]
- Kiessling P, Kuppe C. Spatial multi-omics: Novel tools to study the complexity of cardiovascular diseases. Genome Med. 2024;16(1):14.
[Crossref] [Google Scholar] [PubMed]
- Mourad O, Yee R, Li M, Nunes SS. Modeling heart diseases on a chip: Advantages and future opportunities. Circ Res. 2023;132(4):483-497.
[Crossref] [Google Scholar] [PubMed]
- Liu J, Wang Y. Advances in organ‐on‐a‐chip for the treatment of cardiovascular diseases. MedComm Biomater Appl. 2023;2(4):e63.
- Mistrulli R, Ferrera A, Salerno L, Vannini F, Guida L, Corradetti S, et al. Cardiomyopathy and sudden cardiac death: Bridging clinical practice with cutting-edge research. Biomedicines. 2024;12(7):1602.
[Crossref] [Google Scholar] [PubMed]
Citation: Taleb YS, Memon P, Jalbani A, Al-Mutairi N, Al-Mukhaylid S, Al-Anazi N, et al. (2024). Genotype-phenotype Correlations in Inherited Cardiomyopathies, their Role in Clinical Decision-making and Implications in Personalized Cardiac Medicine in Multi-omics as Well as Disease Modelling Eras. J Clin Exp Pharmacol. 14:431.
Copyright: © 2024 Taleb YS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.