Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 14, Issue 2
Genetic Polymorphisms of Seven Trichoderma Spp Strains, their Potential as Biological Control Agent and Growth Promoter in Tomato
Samuel Baca1, Oswalt R. Jiménez2, Dorian González3, Jorge A. Huete-Pérez3, Rogelio Trabanino1 and Mavir Carolina Avellaneda1*2The Institute for Training, Research and Environmental Development (CIDEA), University of Central America, Nicaragua
3Molecular Biology Center, University of Central America, Nicaragua
Received: 28-Mar-2023, Manuscript No. JPPM-23-20442; Editor assigned: 30-Mar-2023, Pre QC No. JPPM-23-20442(PQ); Reviewed: 14-Apr-2023, QC No. JPPM-23-20442; Revised: 21-Apr-2023, Manuscript No. JPPM-23-20442(R); Published: 28-Apr-2023, DOI: 10.35248/2157-7471.23.14.663
Abstract
Currently, many conventional methods for crop management have been insufficient to cope with the effects of climate change on food production such as droughts, heat, soil nutrient deficiency, and the occurrence of new pathogen strains. Utilizing biological inputs, such as Trichoderma spp, has proved useful in improving crop production and food safety. However, in Central American countries there is not enough expertise to accelerate, in a practical way, its use on a broader scale. Here, we report for the first time the study of a collection of Trichoderma spp strains from Nicaragua and Honduras with the purpose of analyzing their genetic diversity and their potential as biological control agents and growth promoters in tomatoes. Genetic diversity was estimated by sequencing the nuclear ribosomal Internal Transcribed Spacer (ITS) region. After, bifactorial experiments for testing the potential of two strains (TN1C and TC01) and two structures (conidia and microsclerotia) for controlling a harmful strain of Fusarium solani were conducted, followed by bifactorial experiments, considering the same factors, but analyzing their effects as a growth promoter in greenhouse conditions. The DNA sequences amplified from ITS regions (1-F and 4) indicate that there are two species, T. asperellum and T. harzianum, instead of one, consistent with morphological observations. Bayesian and parsimony modeling clustered Trichoderma strains by species providing novel insights about phylogenetic relationships and nucleotide polymorphisms. The strains TN1C and TC01 showed a reduction in the percentage of damage caused by F. solani. In addition, these strains increase the percentage of seed germination, plant height, stem diameter, and the number of leaves in tomato seedlings and plants. Root length and volume increased only in seedlings. Microsclerotia had a better performance with respect to conidia on plant growth development. These results strengthen ongoing research projects and incipient biological control programs oriented to benefit tomato farmers.
Keywords
Biological control; Conidia; DNA sequencing; Microsclerotia; Trichoderma spp
Abbreviations
HA: Hot Air; HW: Hot Water; RH: Relative Humidity; MEA: Malt Extract Agar.
Introduction
The improvement of crop management techniques has been a constant challenge for agricultural scientists since the Green Revolution. Even though the global statistics traced between 1997 to 2017 suggest that there has been significant progress in increasing yields of economically important crops such as maize, rice, soybean and wheat (FAOSTAT, 2019), conventional methods and technologies have not succeeded in coping with recent variable climate events, causing alarm in many food systems [1,2]. The Intergovernmental Panel on Climate Change reported that temperature increases are expected to accelerate to 1.5°C between 2030 and 2052, affecting habitats of around 6% of insects and 8% of plant species [3]. When high temperature coincides with abnormal intervals of rain precipitations, damages and losses on field production are even more profuse. Additionally, under that combination of factors, the dynamics of pest and disease populations change, inverting levels of incidences and promoting the occurrence of more severe strains without any possibility of control because of lacking previous experiences [4,5]. Chemical control by means of using synthetic pesticides has provided a feasible solution for crop protection in economic terms, but its continued and increased use threatens food safety and contributes to creating resistance in pests and pathogens over time [6]. Thus, it is important to identify novel alternatives that fulfil this purpose, but with an integrated crop management vision. In this sense, the development and utilization of biological inputs in agriculture has been identified as one of the eight principles of integrated pest management [7]. These compounds, based on the use of beneficial microorganisms, have received special attention in recent years because its impacts and practicality for small and even large farming systems. The endophytic fungi species from the genus Trichoderma encompasses several species with variable biotechnological applications, being the most important its use as biological agent for the control of a diversity of plant pathogens [8]. In nature, Trichoderma acts as symbiont, colonizing plant root systems and secreting proteins that interact with host genome, this relationship induces significant effects on plant health and production [9]. Numerous studies demonstrate the multiple benefits of inoculating crop plants with Trichoderma spp. strains; among them the induced tolerance to abiotic stresses such as soil salinity, water scarcity, anoxic conditions and heat shock [10,11]. The presence of Trichoderma in roots induces the up regulation of genes and certain pigments expressions that improve photosynthetic capability of plants by activating biochemical pathways that reduce reactive oxygen species to less destructive molecules [12]. Also, this symbiosis optimizes nutrient uptake and consumption by increasing root density and providing tolerance to diverse plant pathogens [13-17]. In the Central American countries, some projects have taken advantage of the existing knowledge concerning Trichoderma to propose practical solutions to many challenges in small-scale agriculture. Currently, seven strains have been suggested for agronomic management of tomato. However, the lack of scientific basis on those proposals has reduced the possibilities of scaling up to larger areas, limiting the impacts on tomato production. Most of the strains produced at laboratory level and used on fields are classified as Trichoderma harzianum, but there is no certainty of the real taxonomic identity of the species using molecular tools. Also, the antagonist level of those strains in the presence of important vegetable pathogens such as Fusarium solani has not been tested before, followed by robust greenhouse experiments that demonstrate the additional effects as growth parameters. The present study aimed to analyze the genetic diversity and phylogenetic relationships among seven Trichoderma spp. strains used in Central America, to evaluate the antagonist level before a F. solani strain and the effects as growth promoter in tomato.
Materials and Methods
Trichoderma strain selection
Seven Trichoderma spp strains: TZ01, TC01, TC02, TN1A, TN1B, TN1C y TN2C from Zamorano University, Honduras were selected for this study, considering their availability and its significance as product in Central America. DNA from eight additional strains: R5BF2M2, TRIIIFIM2, R6CREF4M5, TRIIF3M2, R5KHFIM5A, R5TFIMI, R5IPF2MIB, and MMRI from the Nicaraguan Institute of Agricultural Technology (INTA, Spanish acronym) were included in molecular characterization as well as an isolate of F. solani collected and conserved by the Laboratory of Plant Pathology, Diagnosis and Molecular Research of Zamorano University. This strain of F. solani was stated as outgroup and quality control of the sequences in the phylogenetic tree.
Molecular characterization
Molecular characterization was performed in the Molecular Biology Center of the University of Central America.
DNA extraction and PCR amplification
The DNA of Trichoderma spp and F. solani strains from Zamorano were extracted from pure mycelium cultures, using the Wizard® Promega "Genomic DNA Purification Kit" with modifications of the factory protocol. DNA samples of strains from INTA were kindly provided by that institute. After DNA extracting, quality (260/280) and concentrations (µg/mL) with 320 corrections were estimated using a PerkinElmer Lambda 25 brand spectrophotometer. DNA integrity was measured by electrophoresis in 0.75% agarose gel stained with 0.5 µg/mL of ethidium bromide [18-20]. Primers ITS1-F, ITS4 and ITS4B were used for DNA amplification. PCR reactions were performed using a Phusion High-Fidelity PCR Master Mix with HF Buffer following manufacturer protocol at quarter of reaction (Thermo Fisher Scientific, n.d.-b). The thermocycling conditions were: initial denaturation at 98 ºC for 30 seconds (s), 35 cycles of 98°C for 5 s, 53°C for 30 s and 72°C for 30 s, and a final extension of 72°C for 7 minutes (min). The product band uniformity, integrity and expected size were confirmed in 2% agarose gel stained with ethidium bromide at a concentration of 0.5 µg/ml. Dimers, oligos and any residual component were removed of the reaction by adding 0.4 µL of distilled and autoclaved water, 0.4 µL of rSAP and 0.2 µL of Exo I for each 4 µL of PCR product. The thermal cycler set up consists of 30 min hold at 37°C to activate the enzyme and 15 min at 85°C to denature it [21,22].
Capillary electrophoresis and analysis of sequencing electropherograms
Sequencing samples were prepared for cycle sequencing in 5.5 µL reactions, composed of 0.75 µL of Big Dye Terminator v3.1, 3 µL of distilled and autoclaved water, 0.75 µL of primers, either forward (ITS 1-F) or reverse (ITS 4) and 1 µL of clean PCR product. Reaction conditions were subjected to the manufacturer thermocycling protocol with minor modifications consisting of 30 cycles of 96°C for 10 s, 50°C for 15 s and 60°C for 180 s [22]. Prior to sequencing, the DNA pellet of the previous reaction was precipitated and cleaned using the EDTA protocol for BDT version 3.1 with minor variants [22]. Each PCR product (5.5 µL) was mixed with 1.25 µL of EDTA and 15 µL of 100% ethanol, vortexing at 1,000 revolutions per minute (rpm). Subsequently, the total mixture was transferred to a 1.5 mL microcentrifuge tube and incubates at room temperature for 15 min. Then, it was centrifuged at 18,985 relative centrifugal force (rcf) and 4°C for 15 min, discarding the supernatant and with a spin down at 14,000 rpm for 10 s. Samples were cleaned by adding 15 µL of 70% ethanol and centrifuged in the same way at 18,985 rcf for 15 min, discarding the supernatant and letting it dry for 15 min at room temperature. Finally, 10 µL Hi-Di Formamide was added to the bottom of the tube mixing by lightly pipetting. DNA sequencing was performed using a SeqStudio ABI 3200 machine (Applied Biosystems). In the plate, 10 µL of the previous mix for each sample were added and the run conditions were as manufacturer configuration. Initially, the quality of electropherograms of the sequences were analyzed in Sequencing Analysis Software version 6. Then, sequences were trimmed and detail viewed in the program Sequencing Scanner version 2. Variant Reporter Software version 2 was used to review discrepancies between both, forward and reverse sequences, from the same sample replicates. These three software packages are integrated into SeqStudio ABI 3200 machine. Sequences were aligned in MEGAX creating a unique FASTA sequence per sample [23]. Unique sequences were blasted in the nucleotide database of NCBI (National Center for Biotechnology Information) (https://blast.ncbi.nlm.nih.gov/Blast.cgi), The Barcode of Life Data Systems (BOLD) (http://www.boldsystems.org/index.php/IDS_OpenIdEngine) and TrichOkey (http://www.isth.info/tools/molkey/) in order to identify the species.
Phylogenetic modelling
Files containing DNA sequences previously aligned and edited were converted into*.nex format for phylogenetic analyses. A heuristics searching for best model was conducted by means of Computation of Likelihood Scores and Akaike Information Criterion (AIC) calculations using software JModelTest version 2.1.10 [24,25]. The best model was used to set up parameters for Bayesian analyses, MCMC (Markov Chain Monte Carlo) runs, applying 1,000,000 generations, a frequency of sampling of 100, four runs and chains respectively, all these analyses were performed using software MrBayes version 3.2.7b [26]. Additionally, a bootstrapping with 3,000 pseudo-replicates was performed followed by a simple search with 3,000 repetitions by Parsimony. This analysis was conducted using software POY version 5 [27].
In vivo experiments
In vivo antagonism tests using a F. solani strain: Previously, the seven Trichoderma strains were confronted by means of in vitro dual tests against F. verticilloide, F. delphinoide and F. solani strains (data not shown). After those tests, the most promising strains T. harzianum (TN1C) and T. asperellum (TC01) were chosen to set in vivo evaluations against a F. solani strain which behaved as the most harmful in previous evaluations using tomato plants. For the production of conidia and microsclerotia of Trichoderma spp, a liquid medium was prepared containing: sucrose, yeast extract, KH2PO4, CaCl2 and MgSO4 that was inoculated with the strain from pure culture. The inoculum was stirred at 250 rpm for 72 h for the production of conidia and 120 h for the production of microsclerotia. For the production of conidia of F. solani, the inoculum was prepared in Sabouraud liquid medium, by mixing it with pure culture (168 h old). This mixture was stirred at 250 rpm for 72 h. After, 400 mL of F. solani suspension with a minimum concentration of 9 × 106 conidia mL-1 were applied per each kg of substrate seven days before seedling transplanting. During seedling transplanting, 250 mL of conidia and 250 mL of microsclerotia of strains TN1C and TC01 were applied at a concentration of 1 × 106 conidia mL-1 and 1.3 × 105 microsclerotia mL-1, respectively. The experiment was established in pots with a diameter of 18 cm, a depth of 15 cm and a capacity volume of 2 kg. One seed per pot was sown. The substrate used was composed of soil, compost and rice husks (2:1:1) and sterilized with steam. Fertilizer 18-46-0 (N, P, K) was applied seven days after transplanting and 0-0-60 30 days after (3 g per pot). Weeds were manually removed every seven days. Irrigation was applied with an interval of 24 h to maintain humidity at field capacity. No disease control was conducted. A bi-factorial experiment (2 × 2) was established in a Randomized Complete Block Design (RCBD) with four repetitions and four plants per repetition for a total of 96 experimental units. The treatments evaluated correspond to the combination of two Trichoderma strains: TN1C (T. harzianum) and TC01 (T. asperellum) and two reproductive structures (conidia and microsclerotia), a positive (only F. solani) and a negative (sterile water) control were included. At 41 days after transplanting, the percentage of incidence and severity of diseases were measured.
In vivo growth promoter tests: The experiment was stablished in 50-celled trays, sowing one tomato seed per cell, each tray was considered a repetition. The substrate used was based on soil, compost and rice husk (in a ratio 2:1:1), and sterilized with steam. In this trial, no nutrition or disease control management were performed. Weed control was carried out manually every seven days. Water applications were carried out 2-3 times per day to keep humidity at field capacity. The production of conidia and microsclerotia of both Trichoderma species was conducted in the same way as describe in previous tests. Prior to sowing the seed, 250 mL of the treatments were applied per tray at a concentration of 1 × 106 conidia mL-1 and 1.3 × 105 microsclerotia mL-1, respectively. A bi-factorial experiment (2 × 2) was established in a RCBD with three repetitions for a total of 15 experimental units. The treatments evaluated correspond to the combination of two Trichoderma strains: TN1C (T. harzianum) and TC01 (T. asperellum) and two reproductive structures (conidia and microsclerotia). A negative control (sterile water) was also included. The number of germinated seeds was counted during the first 10 days after sowing. To determine the germination percentage (GP), the following formula was used:
GP=[ n/N ] × 100
Where “n” is the number of normal seedlings and “N” is the number of established seeds. At 27 and 41 days after sowing, the plant height (cm) was measured from the base of the stem to the youngest leaf of the plant; stem diameter (mm) was measured at 1 cm from the base of the plant using a vernier caliper; and number of total leaves of five random seedlings per repetition for a total of 15 seedlings per treatment. At 41 days, the root system of five random seedlings per repetition (15 seedlings per treatment) was evaluated, using the WinRHIZO® program where the root length (cm) and volume (cm3) were estimated.
Statistical analyses: The analyses of the variables disease incidence and severity were merely descriptive. On the other hand, variables: plant height, stem diameter, number of seedling leaves, root length and root volume of plants were analyzed through Analysis of Variance (ANOVA). The percentage of germination, plant height, stem diameter and number of leaves were also analyzed in repeated measures over time, using the program Statistical Analysis System (SAS version 9.4®). For the variables that found significant differences, the means comparisons were conducted by Duncan’s test (α=0.05).
Results
Species identification and phylogenetic relationship among strains
The DNA amplification using ITS1-F and ITS4 primers had better yield and a single band in the ~600–650 bp range. On the other hand, the combination ITS1-F and ITS4B amplified with multiple non-specific bands. This was expected since ITS4B is specific for Basidiomycota fungal samples. KB basecaller generates quality values from 1 to 99 per base and overall sequence, typical high quality mixed bases will have QV 10 to 50 and high quality pure bases will have QV 20 to 50. Only sequences with quality values of 30 (99.9% of confidence) at minimum were accepted in the downstream bioinformatic processes. The minimum overall sequence quality value was 34 (>99.9%) and maximum was 50 (99.999%) in all replicates per sample. Blast results in NCBI nucleotide collection as well as BOLD provided score values higher than 99.9% and E-values of zero at species level identification. In the oligonucleotide barcode program TrichOkey the identification reliability was high for all FASTA sequences submitted, which give a high level of certainty that each of the species was assessed correctly, resulting in two species, Trichoderma asperellum (10 strains) and T. harzianum (five). Most Trichoderma strains from INTA were classified as T. asperellum with exception of MMRI identified as T. harzianum. In contrast, strains from Zamorano were classified in both species. TC01, TC02 and TZ01 were T. asperellum while TN2C, TN1B, TN1C and TN1A were T. harzianum. The differences between these two species were located at 64 different nucleotide (nt) positions. Bayesian parameters for simulations were set up using model F81+G (lset snt=1, rates=gamma) obtained from heuristics searching. Both, Bayesian and Parsimony analyses conducted using haplotypes showed similar results, clustering strains within identified species. The results obtained from phylogenetic analyses are showed in (Figure 1A). Cluster I includes T. asperellum strains: TC01, TC02, R5BF2M2, TRIIIFIM2, R6CREF4M5, TRIIF3M2 and reference for T. asperellum (101201). Four T. asperellum strains: TZ01, R5KHFIM5A, R5TFIMI and R5IPF2MIB did not cluster at this branch due to a thymine (T) deletion at 180 nt and transversion of T to guanine (G) at 438 nt (Figure 1B). Consequently, they are shown as a sub-group within Cluster I with Bayesian posterior probability of 100 and a maximum parsimony bootstrap value of 1. Cluster II encompasses reference strains for T. longibrachiatum (5548) and T. citrinoviride (58853) species, strongly supported (100/0.99). Cluster III gathered T. harzianum strains TN2C, TN1B, TN1C, MMRI, TN1A and reference for T. harzianum species (5544) with Bayesian posterior probability of 100 and a maximum parsimony bootstrap value of 0.99. At this cluster, MMRI showed a deletion of T at 144 nt and TN1C exhibited a transversion of G to cytosine (C) at 576 nt. Finally, reference sample of F. solani, used as outgroup to identify the tree root was clearly classified as cluster IV. References sequences were included to robust phylogenetic inference.
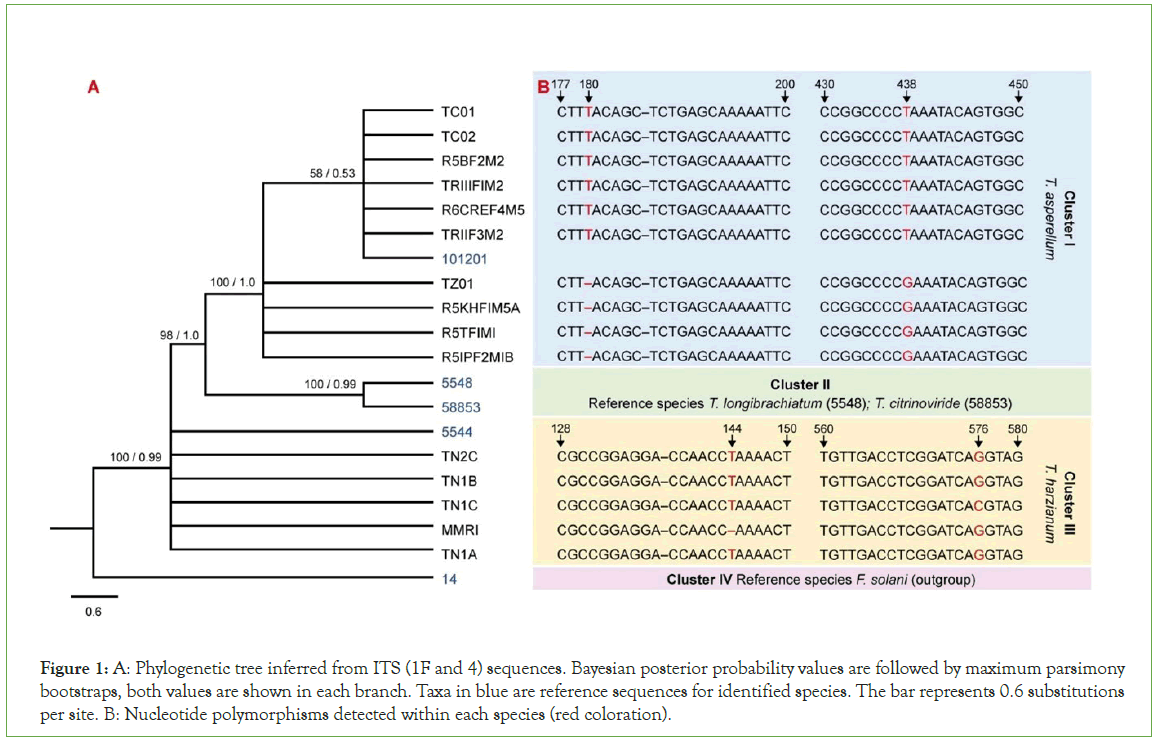
Figure 1: A: Phylogenetic tree inferred from ITS (1F and 4) sequences. Bayesian posterior probability values are followed by maximum parsimony bootstraps, both values are shown in each branch. Taxa in blue are reference sequences for identified species. The bar represents 0.6 substitutions per site. B: Nucleotide polymorphisms detected within each species (red coloration).
In vivo antagonism tests using a F. solani strain
The incidence and severity data obtained at 41 days after transplanting showed that in general the use of Trichoderma strains effectively reduces the damage of wilt disease caused by F. solani in comparison with the positive control. The incidence of the disease in the positive control was 100%, which indicates that there was a successful inoculation. There were differences between Trichoderma strains, but not for reproductive structures. The incidence in Trichoderma treatments was 28.13% (strain TC01 (T. asperellum)) and 53.13% (strain TN1C (T. harzianum)), which would mean a decrease of 71.87% and 46.87% with respect to the positive control. The negative control (without F. solani inoculum) presented an incidence of 37.50%. The positive control presented the highest percentage of damage caused by F. solani in tomato plants with 41.25%, which was different with respect to Trichoderma treatments (7.50% for TC01 (T. asperellum) and 14.38% for TN1C (T. harzianum)) and the negative control 12.50%. The application of TC01 (T. asperellum) decreased the severity of the disease by 33.75% and the application of TN1C (T. harzianum) by 26.87%. A summary of these qualitative results is presented in (Table 1).
| Treatment | Variables | |
|---|---|---|
| Incidence (%) | Severity (%) | |
| TC01 (T. asperellum) | 28.13 | 7.50 |
| TN1C (T. harzianum) | 53.13 | 14.38 |
| Negative control | 37.50 | 12.50 |
| Positive control | 100 | 41.25 |
Table 1: Effect of Trichoderma spp strains TC01 and TNC1 on the incidence and severity of F. solani in tomato plants.
In vivo growth promoter tests
Statistical analysis on the germination percentage showed that there was a significant difference only in the strain factor (P=0.0001) and time of counts (P<0.0001). Although the germination percentage of these seeds was low, probably due to seed aging, it is evidenced that the use of Trichoderma increases the seed germination percentage between 17.60% (TN1C (T. harzianum)) and 18.85% (TC01 (T. asperellum)) with respect to the control (Table 2; Figure 2). The results obtained from growth variables at 27 days after sowing showed that the strains had a significant difference in the three variables. The use of Trichoderma regardless of the strain promotes an increase of plant height (TC01 (T. asperellum) 5.11 cm and TN1C (T. harzianum) 4.88 cm), stem diameter (TC01 (T. asperellum) 2.18 mm and TN1C (T. harzianum) 2.12 mm) and number of leaves (TC01 (T. asperellum) 2.86 and TN1C (T. harzianum) 2.93) (P=0.0001; P=0.0104; P<0.0001, respectively). The reproductive structure showed a significant difference in the variables plant height (P=0.0100) and number of leaves (P=0.0304). There were no significant differences in the species × reproductive structure interaction. The use of microsclerotia (5.30 cm) had a better performance increasing 1.42 cm the seedling height with respect to the control (3.88 cm) and 0.60 cm with respect to the use of conidia (4.70 cm). In the case of leaf production, the use of both conidia (2.80 leaves per seedling) and microsclerotia (3.00 leaves per seedling) increased leaf production between 0.27 and 0.47 leaves per seedling in relation to the control (2.53) (Table 3). The results of the biometric variables, at 41 days after seedling transplanting, showed that the use of Trichoderma has an effect on the growth of the crop in plant height (TC01 (T. asperellum) 34.50 cm and TN1C (T. harzianum) 34.55 cm), stem diameter (TC01 (T. asperellum) 6.21 mm and TN1C (T. harzianum) 6.19 mm) and number of leaves ((T. asperellum) 9.39 and TN1C (T. harzianum) 9.43 leaves per plant) compared to controls. The reproductive structure has a minor influence, showing a significant difference only in the plant height (P=0.0002), being the conidia (34.98 cm) statistically superior with respect to the microsclerotia (34.07 cm) and the control (32.69 cm) (Table 4). The interactions species × reproductive structures over time showed significant differences in plant height (P<0.0001) and diameter (P=0.0018). In these variables, the treatments began to differ after 13 days showing that the interactions that involve conidia as a reproductive structure (T. harzianum+conidia and T. asperellum+conidia) have a better performance compared to microsclerotia in time. The results from the variables of the root system at 28 days after seedling transplanting showed significant differences for strains and reproductive structure in all variables, but not so for the interaction species × reproductive structure. In plants 41 days after seedling transplanting, there was no significant difference in any variable, that is, the application of these Trichoderma strains and its reproductive structures did not have an effect on tomato root development. The use of Trichoderma promotes root growth by increasing the length of roots by 70.95% (TC01, T. asperellum, 278.27 cm) and 68.87% (TN1C, T. harzianum, 274.88 cm) and root volume by 60.00% (TC01, T. asperellum, 0.32 cm3) and 45.00% (TN1C, T. harzianum, 0.29 cm3) with respect to the control (162.77 cm of root length and 0.20 cm3 of root volume) (Table 5; Figure 3). Regarding the reproductive structure, the effect of microsclerotia is statistically different in root length (323.71 cm) and volume of roots (0.33 cm3) increasing by 41.08% and 22.22%, respectively compared to conidia (length 229.44 cm and volume 0.27 cm3) and with the control (length 162.77 cm and volume 0.20 cm3) (Table 5).
| Treatments | Germination (%)z* |
|---|---|
| TN1C (T. harzianum) | 41.77 ± 2.87 a |
| TC01 (T. asperellum) | 43.02 ± 4.13 a |
| Control | 24.17 ± 3.56 b |
| P value (model) | <0.0001 |
| Pr>Strain (S) | 0.0001 |
| Pr>Reproductive structure (RE) | 0.1658 |
| Pr>Time (T) | <0.0001 |
| Pr>Interaction (S × RE × T) | 0.5914 |
| R2 | 0.6440 |
| CV (%) | 36.7628 |
Note: z*Means in columns with the same letter are not significantly different at the significance level of P ≤ 0.05 according to Duncan's test.
Table 2: Effect of two strains of Trichoderma (TN1C and TC01) and two reproductive structures (conidia and microsclerotia) on the germination of tomato seeds.
| Variables | |||
|---|---|---|---|
| Treatment | Plant height (cm) z* | Stem diameter (mm) z* | Number of leaves z* |
| Strain | |||
| TN1C (T. harzianum) | 4.88 ± 0.19 a | 2.12 ± 0.06 a | 2.93 ± 0.08 a |
| TC01 (T. asperellum) | 5.11 ± 0.17 a | 2.18 ± 0.04 a | 2.86 ± 0.06 a |
| Control | 3.88 ± 0.20 b | 1.70 ± 0.06 b | 2.53 ± 0.13 b |
| Reproductive structure | |||
| Conidia | 4.70 ± 0.15 b | 2.07 ± 0.04 a | 2.80 ± 0.07 a |
| Microesclerotia | 5.30 ± 0.19 a | 2.23 ± 0.06 a | 3.00 ± 0.06 a |
| Control | 3.88 ± 0.20 c | 1.70 ± 0.06 a | 2.53 ± 0.13 b |
| Pr>Strain (S) | 0.0001 | 0.0104 | <0.0001 |
| Pr>Reproductive structure | 0.0100 | 0.0658 | 0.0304 |
Note: z*Means in columns with the same letter are not significantly different at the significance level of P≤ 0.05 according to Duncan's test.
Table 3: Effects of two strains and two reproductive structures of Trichoderma spp on plant height stem diameter and number of leaves in tomato plants 27 days after sowing.
| Treatment | Variables | ||
|---|---|---|---|
| Plant height (cm)z* | Stem diameter (mm)z* | Number of leavesz* | |
| Strain | |||
| TN1C (T. harzianum) | 34.55 ± 1.90 a | 6.19 ± 0.23 a | 9.43 ± 0.50 a |
| TC01 (T. asperellum) | 34.50 ± 1.89 a | 6.21 ± 0.23 a | 9.39 ± 0.50 a |
| Control + | 32.71 ± 2.65 b | 5.94 ± 0.32 b | 9.28 ± 0.71 ab |
| Control - | 32.67 ± 2.54 b | 5.80 ± 0.30 b | 9.00 ± 0.68 b |
| Reproductive structure | |||
| Conidia | 34.98 ± 1.92 a | 6.27 ± 0.23 a | 9.44 ± 0.50 a |
| Microesclerotia | 34.07 ± 1.86 b | 6.14 ± 0.23 a | 9.39 ± 0.50 a |
| Control + | 32.69 ± 1.83 c | 5.87 ± 0.22 a | 9.14 ± 0.49 a |
| Pr>Strain (S) | <0.0001 | <0.0001 | 0.0413 |
| Pr>Reproductive structure | 0.0002 | 0.0504 | 0.7182 |
Note: z*Means in columns with the same letter are not significantly different at the significance level of P≤ 0.05 according to Duncan's test
Table 4: Effects of two strains and two reproductive structures of Trichoderma spp on plant height stem diameter and number of leaves in tomato plants 41 days after sowing
| Treatment | Variables | |
|---|---|---|
| Root length (cm)z* | Root volume (cm3)z* | |
| Strain | ||
| TN1C (T. harzianum) | 274.88 ± 18.54 a | 0.29 ± 0.01 a |
| TC01 (T. asperellum) | 278.27 ± 16.43 a | 0.32 ± 0.01 a |
| Control | 162.77 ± 16.58 b | 0.20 ± 0.00 b |
| Reproductive structure | ||
| Conidia | 229.44 ± 12.70 b | 0.27 ± 0.01 b |
| Microesclerotia | 323.71 ± 17.30 a | 0.33 ± 0.02 a |
| Control | 162.77 ± 16.58 c | 0.20 ± 0.00 c |
| Pr>Strain (S) | <0.0001 | 0.0003 |
| Pr>Reproductive structure | <0.0001 | 0.0051 |
Note: z*Means in columns with the same letter are not significantly different at the significance level of P≤ 0.05 according to Duncan's test
Table 5: Effects of two strains and two reproductive structures of Trichoderma spp on root length (cm) and root volume (cm3) in tomato plants 28 days after sowing.
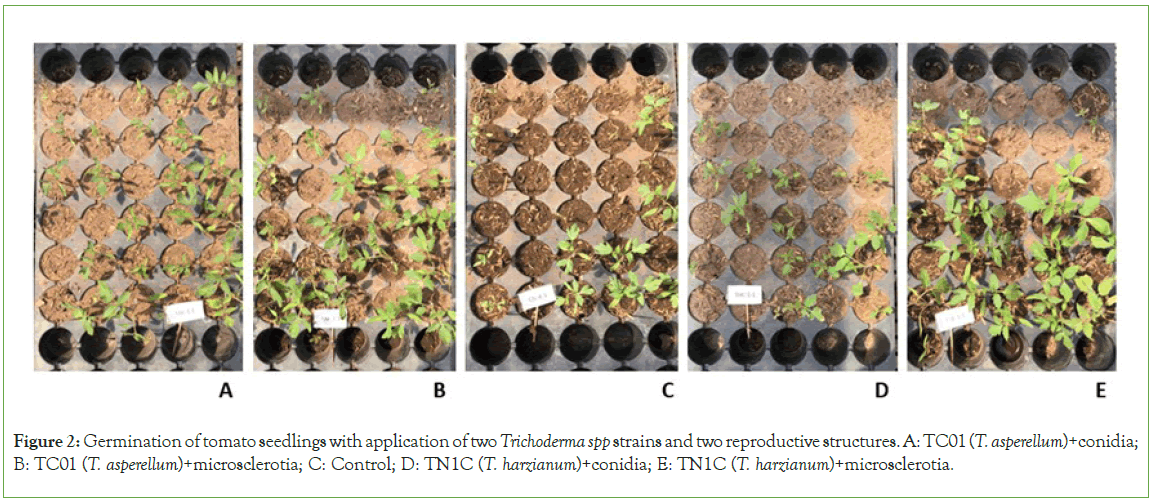
Figure 2: Germination of tomato seedlings with application of two Trichoderma spp strains and two reproductive structures. A: TC01 (T. asperellum)+conidia; B: TC01 (T. asperellum)+microsclerotia; C: Control; D: TN1C (T. harzianum)+conidia; E: TN1C (T. harzianum)+microsclerotia.
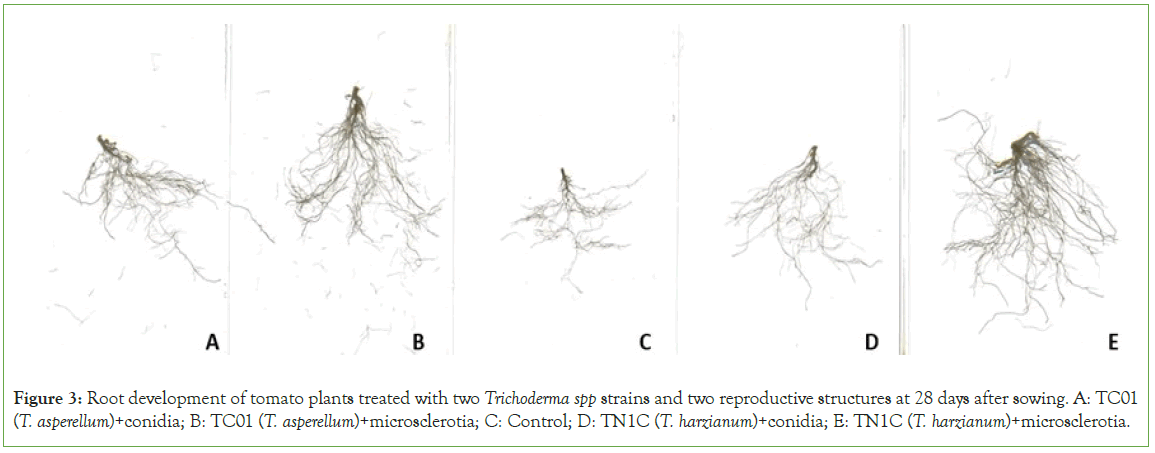
Figure 3: Root development of tomato plants treated with two Trichoderma spp strains and two reproductive structures at 28 days after sowing. A: TC01 (T. asperellum)+conidia; B: TC01 (T. asperellum)+microsclerotia; C: Control; D: TN1C (T. harzianum)+conidia; E: TN1C (T. harzianum)+microsclerotia.
Discussion
A complete understanding of the taxonomical identity, molecular polymorphisms, antagonism level, and potential as a plant growth promoter of a collection of Trichoderma spp strains is essential for any biological control program. Here, we provided key information that could strengthen current strategies at the same time that optimizes inputs use in tomato cultivation.
There are two different Trichoderma species with polymorphisms among strains
Most previous efforts have been focused on describing genetic and phylogenetic characteristics of novel Trichoderma isolates from different origins and their potential as biocontrol agent in a broad number of pathogen species such as Alternaria spp., Botryodiplodia theobromae, Curvularia sp., Macrophomina spp., Mucor racemosus, Candida albicans, Plasmopara viticola, Pseuperonospo cubensis, Rhizopus spp., Sphaerotheca fusca, Rhizoctonia spp., Aspergillus spp., Botrytis spp., Colletotrichum spp., Pseudomonas syringae, Sclerotinia spp., Xanthomonas campestris;, Fusarium spp., Pythium spp., Phytophthora spp., Sclerotium rolfsii and Rhizoctonia [28-38]. But, there are no references of studies that describe the genetic diversity and polymorphisms of strains under production and promotion in tomato crop. According to DNA sequence analyses, strains were classified into two species T. asperellum and T. harzianum, correcting previous perception of having only one species. Indeed, some reports indicate that several isolates of genus Trichoderma have been initially misclassified because of their similarity, scarcity of morphological characters, and lacking molecular tools needed to confirm taxonomic identity [39]. Because T. harzianun is the most abundant species in diverse niches, it is very common to attribute any novel isolate to that species. Usually, DNA sequences are compared to reference databases in order to achieve taxonomic identification, a study of the assessment of the main public repositories of DNA Barcode sequences, BOLD and GeneBank, accuracy and reliability outlined that both databases performed comparably for fungi identification, and described BOLD as a curation tool that also stores sequences and GenBank just as a sequence repository with basic quality checks [40]. With the purpose of having a high level species identification confiability, we used a third database repository, TrichOkey, which is a method for molecular identification of Trichoderma at the genus and species levels, using a combination of several genus specific hallmarks and species clades specifically allocated within the internal transcribed spacer 1 and 2 (ITS1 and 2) sequences of rDNA repeat. All top hits from blasts in GenBank and BOLD provided an E-value of 0.0, with expected identity scores higher than 99.9% and were an exact match with TrichOkey results. Hence, DNA sequences obtained using ITS1-F and ITS4 primers were capable of disaggregating strains into two species with high level of confidence (>99.9%) without the need of sampling more genome regions for this purpose. However, it would probably be necessary to analyze at least two additional gene regions to detect more nucleotide polymorphisms for each strain, for traceability purposes or to explore more secondary fungal DNA barcodes such as the stated optimal for secondary DNA barcoding in Ascomycota, DNA topoisomerase I (TOPI) and phosphoglycerate kinase (PGK) [40,41]. All these options are promising for fingerprinting all the Trichoderma strains. Phylogenetic analyses suggested a consensus tree (considering both approaches, Bayesian and Parsimony) desegregating strains into two main clusters according to species. This result was expected considering the reduced number of taxa and species. There was no apparent correlation between origin and phylogenetic structure as reported in other studies where geographic origin and host crop influence the genetic structure of Trichoderma populations. On the other hand, it was interesting that our results confirmed that strain TZ01 was clearly different from strains TC01 and TC02, something that has been suggested by researchers at Zamorano University based on morphological information (personal observation). Within cluster I, T. asperellum strains were divided into two sub-clusters, differences are due to a T deletion at 180 nt and a transversion (T-G) at 438 nt. As these two nucleotide changes were consistent in two groups of T. asperellum strains, phylogenetic algorithms conducted in both approaches divided strains into two sub-clusters. On the contrary, in cluster III, strains TN1C and MMR1 showed two separate changes in nucleotide sequences, a T deletion at 144 nt and a transversion (G-C) at 576 nt, respectively. Although, these variations were not significant for sub-branching those taxa during phylogenetic analyses. However, these polymorphisms are useful for strain barcoding. These kind of mutations are reported as the main cause of genetic differentiation within Trichoderma species, affecting in some cases gene functionality and exposing sub-clustering in phylogenetic analyses under diverse approaches [42,43]. It was interesting that during the growth of the strains in PDA media, we observed variations in morphology that suggest that more than one strain could be present in the pure culture. Although it was not confirmed in the present study, it is something to consider in future research. This would make a lot of sense if we consider the complex relationships between microorganisms and plants, as studied in sugarcane where the community plays a key role in the success of the symbiosis. This could change the perception of the “genetic purity of the strains” in the future and adopt the concept of “community-based collections” instead [44,45].
Trichoderma spp reduces incidence and severity of F. solani on tomato plants
The fungus F. solani, causative agent of the wilt disease, is one of the most harmful plant pathogens around the world, acting in some cases in complex with other pathogens, for instance, Pythium spp and Rhizoctonia spp in the damping-off disease in vegetable seedlings. In tomato, wilt disease causes important losses that could be estimated between 20% and 50% depending on environmental conditions, variety and pathogen’s strain. The identification of biological alternatives that control F. solani is of great importance for the improvement of tomato productivity, its safety and the reduction of production costs, something important under current global context. In this study, the results of incidence and severity of F. solani, considering the positive control, demonstrated that Trichoderma spp is an effective alternative for reducing the damage caused by this pathogen. The endogenous presence of F. solani (37.50%) in the positive control is not surprising since it is a seed-borne pathogen frequently found in seeds [46-48]. However, despite of this, the supply of Trichoderma spp improves the health of plants expose to high concentration of inoculum. If we consider another scenario where there is no F. solani inoculum on the field, only the one present in the seed, even so, strain TC01 (T. asperellum) seems to suppress the incidence of the disease by 9.37% and its severity by 5%, improving the performance of infected seeds in nurseries. Indeed, Trichoderma antagonistic effects, such as substrate competition, mycoparasitism, and antifungal antibiotic production, are useful to inhibit the mycelial growth of pathogens [49]. Some hydrolytic enzymes and metabolites could play a key role in controlling Fusarium wilt. Sundaramoorthy and Balabaskar (2013) confirmed that under in vitro conditions the isolate ANR-1 (T. harzianum) was found to effectively inhibit the radial mycelial growth of this pathogen. In the meantime, under greenhouse conditions, this isolate exhibited the least disease incidence (by 15.33%) in the experiment. It is suggested that Trichoderma is able to trigger a long-lasting up-regulation of the salicylic acid pathway even without any pathogen infection, probably stimulating a priming mechanism in the plant [50]. Also, it has been observed the Trichoderma can induce genes involved in the jasmonic and ethylene transduction pathways confirmed by microarray and qRT- PCR analyses, suggesting a transitory increment of plant defense [51].
The use of Trichoderma spp improves the seed germination and growth of tomato plants
Although the physiological quality of the seed lot used in this study was very low, the use of these seeds in these experiments was an opportunity to test the benefits of using Trichoderma spp. It was impressive how Trichoderma spp, regardless of the strain, almost doubled the germination of tomato seeds compared with the control. This demonstrates the potential of these strains used as biological seed protectants, which could provide added value to seed inputs that are often very scarce for agriculture based on less use of inputs. Some studies suggest that Trichoderma spp could modify mRNA levels of 45 genes, 41 in roots and 4 in leaves promoting more growth in tomato seedlings expressed as an increasing in plant height, stem diameter and number of leaves, root length, dry matter parameters [52,53]. In our experiment, even at 27 days after sowing, the variables plant height, stem diameter and number of leaves was still higher in plants inoculated with Trichoderma spp compared to the control, showing that plant growth is effectively promoted beyond germination period. This same trend was observed when analyzing the same variables at 41 days after sowing. In addition of creating a biological protection against pathogens during seed germination, Trichoderma spp establishes an effective symbiosis with tomato plants, steadily promoting plant growth due to the stimulation of the secretion of growth hormones. In this respect, Chowdappa et al. (2013) found that the use of T. harzianum in tomato seedlings increased the levels of indoleacetic acid by 54.34% and gibberellic acid by 67.59% in the root tissue. That study reported increases in shoot length (32.04%), leaf area (62.68%) and fresh weight of shoots (28.87%) in tomato plants. Similar results were obtained by Nzanza et al., (2011) who evaluated the performance of tomato seedlings supplied with T. harzianum at sowing stage or two weeks after sowing during two years. In that study, the application of T. harzianum at both times increased plant height of tomato in 63.41% and 74.29% respectively in 2008 and by 33.67% and 35.65% in 2009, respectively. Other authors report increases in the growth rate of tomato, maize, tobacco, radish [54], beans, cucumber, pepper and certain ornamental species as a result of the application of Trichoderma [55-59]. Although not addressed in depth in this study, it is known that Trichoderma spp can induce an improvement in photosynthetic capacities in plants [60], promoting greater generation of carbohydrates and therefore increased yields of many crops. These benefits can be further enhanced when added to other management actions that improve the availability of nutrients in the soil and the use of sunlight. The supply of Trichoderma spp influenced root growth only in plants 28 days after sowing. The improvement expressed as root length and root volume was similar for both strains, but significantly superior to the control. In average, Trichoderma spp strains improve root length by 41.15% and root volume by 34.43% compared with the control. Nzanza et al. (2011) reported that tomato seedlings inoculated with T. harzianum increased plant root length by 23.20% when T. harzianum was inoculated at planting time and 39.46% when it was applied two weeks after planting obtained similar results. In another experiment, Chowdappa et al. (2013) reported increases of 38.53% in root length and 36.21% in fresh weight of tomato root due to the use of T. harzianum compared to the control. Other authors report average increases of 66% in shoots and roots of sweet corn as well as increases of 75% in root length and 100% in root volume in cucumber. The application of Trichoderma as a seed treatment colonizes the roots of the seedlings and induces their growth and development, which allows them to reach greater depths of soil, improving the dynamics with the microbiota, tolerance to drought, the performance in compacted soils and the yields [61-63]. However, the host plant, pH, temperature and other microorganisms present in the rhizosphere could influence the growth and development of these populations [64]. For example, Nzanza et al. (2011) reported 90% colonization by T. harzianum in tomato seedlings at nursery and 85% in open field conditions. Thus, it is important together with Trichoderma to provide a proper crop management that improve the performance of the binomial Trichoderma–plant, such as incorporation of organic matter to the soil, crop/varieties rotations, and minimum soil tillage [65-70]. The response of tomato plants to the application of Trichoderma was different between the seedling stage and the stages after seedling transplanting. In the early stages of seedlings, Trichoderma directly influenced the increase in root length and volume, an effect that did not occur during the productive stages. This behavior agrees with the results obtained by Nzanza et al. (2012) who evaluated the effect of T. harzianum on the dry weight of tomato roots in the open field without presenting significant differences with respect to the control in two productive cycles [71-78]. However, Nzanza et al. (2011) previously found an average increase of 53.44% of the dry weight of seedlings roots under greenhouse conditions. This behavior is because plants at early stages of development that is during seed germination and seedling growth, they experience the highest growth rates in their life cycle, and then Trichoderma is able to influence significantly the hormone machinery producing the results expressed here and in other studies. The efficiency of the reproductive structure was different depending on the experiments. In the plants that were transplanted, the conidia stood out statistically over the microsclerotia in the interaction over time, as the independent factor. This is contrary to what happened in tomato seedlings at 28 days in which the microsclerotia had a better performance with respect to conidia at plant height. This performance could be due to the effect of environmental factors, in time and space. The temperature and relative humidity values recorded during both experiments (not shown) presented values above those registered in the antagonism experiment (in vivo) under greenhouse conditions. This would be reflected in an increase in the evaporation rate. Then, the soil moisture would be affected, creating stress conditions for both the plant and the Trichoderma strains, influencing the performance of the reproductive structures. In this case, the microsclerotia showed a better behavior under conditions of water stress compared to the conidia, which agrees with the description by Coley-Smith and Cooke (1971).
Conclusion
This first approach provided important insights about genetic diversity, genetic structure and nucleotide polymorphisms among fifteen Trichoderma strains which may be of value for biological control programs in Nicaragua and Honduras in order to detect duplicates in the collections, purity testing or traceability. On the other hand, the use of strains TN1C and TC01 effectively reduced the incidence and severity of F. solani, a harmful tomato pathogen which produced important economic losses around the world. Also, these strains influenced a positive effect on the growth and development of tomato plants, improving the germination of seeds, and increasing plant height, stem diameter and number of leaves in tomato seedlings and plants. Root length and volume increased only in seedlings. The evaluation of the reproductive structures only showed a better performance at the level of seedlings, being the microsclerotia the one with the best performance than the conidia. These results motivate the use of Trichoderma as a key input in tomato nurseries for improving the health and yield of plants. According to climate scenarios, the superior performance of microsclerotia could suggest its use considering the stressing conditions where tomato seedlings could be planted. All these results provided crucial information on the Trichoderma spp as a biological control agent, with practical and economic implications for tomato cultivation.
Declaration
CRediT authorship contribution statement
Samuel Baca: Conceptualization, Methodology, Formal analysis, Investigation, Writing–Original Draft. Oswalt R. Jiménez: Methodology, Formal analysis, Writing–Original Draft, Supervision. Dorian González: Methodology, Formal analysis, Writing–Original Draft, Supervision on molecular methods. Jorge A. Huete-Pérez: Writing–Review and Editing. Rogelio Trabanino: Writing–Review and Editing, Supervision. M. Carolina Avellaneda: Writing–Review and Editing, Supervision.
Competing interest
Authors express non-competing interests in this study.
Acknowledgement
We want to express to our gratitude to Rikolto in Latin America (Nicaragua) for funding paper publication. We thank Andrea Zamora from INTA-Nicaragua for providing in 2019 some DNA samples of Trichoderma spp. strains for this study. We express our gratitude to Yenssy M. Hernández from University of Central America for designing the art of the phylogenetic tree and Sayda Guzmán from Zamorano for helping during strain laboratory culture.
References
- Ray DK, West PC, Clark M, Gerber JS, Prishchepov AV, Chatterjee S. Climate change has likely already affected global food production. PloS one. 2019;14(5):e0217148.
[Crossref] [Google Scholar] [PubMed]
- Solaymani S. Impacts of climate change on food security and agriculture sector in Malaysia. Envi, Devel Sust. 2018;20(4):1575-1596.
- Hoegh-Guldberg O, Jacob D, Taylor M, Guillén Bolaños T, Bindi M, Brown S, et al. The human imperative of stabilizing global climate change at 1.5 C. Science. 2019;365(6459):eaaw6974.
[Crossref] [Google Scholar] [PubMed]
- Jones RA. Future scenarios for plant virus pathogens as climate change progresses. Adv Virus Res. 2016;95:87-147.
[Crossref] [Google Scholar] [PubMed]
- Jones RA, Barbetti MJ. Influence of climate change on plant disease infections and epidemics caused by viruses and bacteria. CABI Rev. 2012;133.
- Popp J, Pet o K, Nagy J. Pesticide productivity and food security. A review. Agron Sustain. 2013;33(1).
- Barzman M, Bàrberi P, Birch AN, Boonekamp P, Dachbrodt-Saaydeh S, Graf B, et al. Eight principles of integrated pest management. Agron Sustain Dev. 2015;35:1199-1215.
- Harman GE. Trichoderma—not just for biocontrol anymore. Phytoparasitica. 2011;39(2):103-108.
- Harman GE, Herrera-Estrella AH, Horwitz BA, Lorito M. Trichoderma–from basic biology to biotechnology. Microbiol. 2012;158(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Brotman Y, Landau U, Cuadros-Inostroza A, Takayuki T, Fernie AR, Chet I, et al. Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 2013;9(3):e1003221.
[Crossref] [Google Scholar] [PubMed]
- Zaidi NW, Dar MH, Singh S, Singh US. Trichoderma species as abiotic stress relievers in plants. In Biotechnol and biol tricho. 2014;515-525.
- Harman GE, Doni F, Khadka RB, Uphoff N. Endophytic strains of Trichoderma increase plants’ photosynthetic capability. J Appl Microbiol. 2021;130(2):529-546.
[Crossref] [Google Scholar] [PubMed]
- Martínez-Medina A, Roldán A, Pascual JA. Interaction between arbuscular mycorrhizal fungi and Trichoderma harzianum under conventional and low input fertilization field condition in melon crops: growth response and Fusarium wilt biocontrol. Appl Soil Ecol. 2011;47(2):98-105.
- Bunbury-Blanchette AL, Walker AK. Trichoderma species show biocontrol potential in dual culture and greenhouse bioassays against Fusarium basal rot of onion. Biol Control. 2019;130:127-135.
- Ghazanfar MU, Raza M, Raza W, Qamar MI. Trichoderma as potential biocontrol agent, its exploitation in agriculture: a review. Plant Prot. 2018;2(3).
- Roberts DP, Maul JE, McKenna LF, Emche SE, Meyer SL, Collins RT, et al. Selection of genetically diverse Trichoderma spp. isolates for suppression of Phytophthora capsici on bell pepper. Can J Microbiol. 2010;56(10):864-873.
[Crossref] [Google Scholar] [PubMed]
- Singh A, Shukla N, Kabadwal BC, Tewari AK, Kumar J. Review on plant-Trichoderma-pathogen interaction. Int J Curr Microbiol Appl Sci. 2018;7(2):2382-2397.
- Pertierra LR, Hughes KA, Vega GC, Olalla-Tárraga MÁ. High resolution spatial mapping of human footprint across Antarctica and its implications for the strategic conservation of avifauna. PloS one. 2017;12(1):e0168280.
[Crossref] [Google Scholar] [PubMed]
- Lucena-Aguilar G, Sánchez-López AM, Barberán-Aceituno C, Carrillo-Avila JA, López-Guerrero JA, Aguilar-Quesada R. DNA source selection for downstream applications based on DNA quality indicators analysis. Biopreserv Biobank. 2016;14(4):26470.
[Crossref] [Google Scholar] [PubMed]
- Sambrook J, Russell DW. Detection of DNA in agarose gels. Mol Clon, A Laboratory Manual,(3rd Ed.) Cold Spring Harbor Laboratory Press, New York. 2001:5-14.
[Crossref] [Google Scholar] [PubMed]
- Toju H, Tanabe AS, Yamamoto S, Sato H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PloS one. 2012;7(7):e40863.
[Crossref] [Google Scholar] [PubMed]
- Climate change has likely already affected global food production
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evel. 2018;35(6):1547.
[Crossref] [Google Scholar] [PubMed]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods. 2012;9(8):772.
[Crossref] [Google Scholar] [PubMed]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696-704.
[Crossref] [Google Scholar] [PubMed]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539-542.
[Crossref] [Google Scholar] [PubMed]
- Wheeler WC, Lucaroni N, Hong L, Crowley LM, Varón A. POY version 5: phylogenetic analysis using dynamic homologies under multiple optimality criteria. Cladistics. 2015;31(2):189-196.
[Crossref] [Google Scholar] [PubMed]
- Błaszczyk L, Popiel D, Chełkowski J, Koczyk G, Samuels GJ, Sobieralski K, et al. Species diversity of Trichoderma in Poland. J Appl Genet. 2011;52:233-243 .
[Crossref] [Google Scholar] [PubMed]
- Druzhinina IS, Kopchinskiy AG, Kubicek CP. The first 100 Trichoderma species characterized by molecular data. Mycosci. 2006;47(2):55-64.
- Druzhinina IS, Komoń-Zelazowska M, Ismaiel A, Jaklitsch W, Mullaw T, et al. Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet Biol. 2012;49(5):358-368.
[Crossref] [Google Scholar] [PubMed]
- Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP. An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol. 2005;42(10):813-28.
[Crossref] [Google Scholar] [PubMed]
- Feitosa YB, Cruz-Magalhães V, Argolo-Filho RC, de Souza JT, Loguercio LL. Characterization of genetic diversity on tropical Trichoderma germplasm by sequencing of rRNA internal transcribed spacers. BMC Res Notes. 2019;12:1-6.
[Crossref] [Google Scholar] [PubMed]
- Hermosa MR, Keck E, Chamorro I, Rubio B, Sanz L, Vizcaíno JA, et al. Genetic diversity shown in Trichoderma biocontrol isolates. Mycol Res. 2004;108(8):897-906.
[Crossref] [Google Scholar] [PubMed]
- Kamala T, Devi SI, Sharma KC, Kennedy K. Phylogeny and taxonomical investigation of Trichoderma spp. from Indian region of Indo-Burma biodiversity hot spot region with special reference to Manipur. Biomed Res Int. 2015;2015.
[Crossref] [Google Scholar] [PubMed]
- Zhang CL, Druzhinina IS, Kubicek CP, Xu T. Trichoderma biodiversity in China: evidence for a North to South distribution of species in East Asia. FEMS microbiol let. 2005;251(2):251-257.
[Crossref] [Google Scholar] [PubMed]
- Filizola PR, Luna MA, de Souza AF, Coelho IL, Laranjeira D, Campos-Takaki GM. Biodiversity and phylogeny of novel Trichoderma isolates from mangrove sediments and potential of biocontrol against Fusarium strains. Microbial cell factories. 2019;18(1):1-4.
[Crossref] [Google Scholar] [PubMed]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2(1):43-56.
[Crossref] [Google Scholar] [PubMed]
- Wu Q, Sun R, Ni M, Yu J, Li Y, Yu C, et al. Identification of a novel fungus, Trichoderma asperellum GDFS1009, and comprehensive evaluation of its biocontrol efficacy. PloS one. 2017;12(6):e0179957.
[Crossref] [Google Scholar] [PubMed]
- Schuster A, Schmoll M. Biology and biotechnology of Trichoderma. Appl Microbiol Biotechnol. 2010;87:787-799.
[Crossref] [Google Scholar] [PubMed]
- Meiklejohn KA, Damaso N, Robertson JM. Assessment of BOLD and GenBank–Their accuracy and reliability for the identification of biological materials. PloS one. 2019;14(6):e0217084.
[Crossref] [Google Scholar] [PubMed]
- Stielow JB, Levesque CA, Seifert KA, Meyer W, Irinyi L, Smits D, et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Pers Mol Phylogeny Evol Fungi. 2015;35(1):242-263.
- Rai S, Kashyap PL, Kumar S, Srivastava AK, Ramteke PW. Identification, characterization and phylogenetic analysis of antifungal Trichoderma from tomato rhizosphere. Springerplus. 2016;5(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Skoneczny D, Oskiera M, Szczech M, Bartoszewski G. Genetic diversity of Trichoderma atroviride strains collected in Poland and identification of loci useful in detection of within-species diversity. Fol microbiol. 2015;60:297-307.
[Crossref] [Google Scholar] [PubMed]
- Armanhi JS, de Souza RS, de Araújo LM, Okura VK, Mieczkowski P, Imperial J, et al. Multiplex amplicon sequencing for microbe identification in community-based culture collections. Sci Rep. 2016;6(1):29543.
[Crossref] [Google Scholar] [PubMed]
- Armanhi JS, De Souza RS, Damasceno ND, De Araujo LM, Imperial J, Arruda P. A community-based culture collection for targeting novel plant growth-promoting bacteria from the sugarcane microbiome. Front Plant Sci. 2018;8:2191.
[Crossref] [Google Scholar] [PubMed]
- Nzanza B, Marais D, Soundy P. Response of tomato (Solanum lycopersicum L.) to nursery inoculation with Trichoderma harzianum and arbuscular mycorrhizal fungi under field conditions. Acta Agricul Scan, Sec B Soil & Plant Sci. 2012;62(3):20915.
- Al-Askar AA, Ghoneem KM, Rashad YM, Abdulkhair WM, Hafez EE, Shabana YM, et al. Occurrence and distribution of tomato seed-borne mycoflora in Saudi Arabia and its correlation with the climatic variables. Microb Biotechnol. 2014;7(6):556-569.
[Crossref] [Google Scholar] [PubMed]
- Mehedi I, Sultana A, Raju MA. Control of seed borne fungi on tomato seeds and their management by botanical extracts. Res Agric Livest Fish. 2016;3(3):403-410.
- Li Y, Sun R, Yu J, Saravanakumar K, Chen J. Antagonistic and biocontrol potential of Trichoderma asperellum ZJSX5003 against the maize stalk rot pathogen Fusarium graminearum. Indian J Microbiol. 2016;56:318-327.
[Crossref] [Google Scholar] [PubMed]
- Tucci M, Ruocco M, De Masi L, De Palma M, Lorito M. The beneficial effect of Trichoderma spp. on tomato is modulated by the plant genotype. Mol plant pathol. 2011;12(4):341-354.
[Crossref] [Google Scholar] [PubMed]
- Moreno CA, Castillo F, González A, Bernal D, Jaimes Y, Chaparro M, et al. Biological and molecular characterization of the response of tomato plants treated with Trichoderma koningiopsis. Physiol Mol Plant Pathol. 2009;74(2):111-120.
- Chowdappa P, Kumar SM, Lakshmi MJ, Upreti KK. Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biocontrol Sci. 2013;65(1):109-117.
- Nzanza B, Marais D, Soundy P. Tomato (Solanum lycopersicum L.) seedling growth and development as influenced by Trichoderma harzianum and arbuscular mycorrhizal fungi.
- Windham MT. A mechanism for increased plant growth induced by Trichoderma spp. Phytopathol. 1986;76:518-521.
- Mayo S, Gutierrez S, Malmierca MG, Lorenzana A, Campelo MP, Hermosa R, et al. Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front Plant Sci. 2015;6:685.
[Crossref] [Google Scholar] [PubMed]
- Pereira JL, Queiroz RM, Charneau SO, Felix CR, Ricart CA, da Silva FL, et al. Analysis of Phaseolus vulgaris response to its association with Trichoderma harzianum (ALL-42) in the presence or absence of the phytopathogenic fungi Rhizoctonia solani and Fusarium solani. Plos one. 2014;9(5):e98234.
[Crossref] [Google Scholar] [PubMed]
- Mei LI, Hua LI, SU XL, Ying TI, HUANG WK, Jie ME, et al. The effects of Trichoderma on preventing cucumber fusarium wilt and regulating cucumber physiology. J Integr Agric. 2019;18(3):607-617.
- Yedidia I, Srivastva AK, Kapulnik Y, Chet I. Effect of Trichoderma harzianum on microelement concentrations and increased growth of cucumber plants. Plant and soil. 2001;235:235-242.
- Chang YC, Chang YC, Baker R, Kleifeld O, Chet I. Increased growth of plants in the presence of the biological control agent Trichoderma harzianum. Plant Dis. 1986;70(2):145-148.
- Harman GE. Development and benefits of rhizosphere competent fungi for biological control of plant pathogens. J Plant Nutr. 1992;15(6-7):835-843.
- Bailey BA, Lumsden RD. Gliocladium on plant growth and resistance to pathogens. Trichoderma and gliocladium: enzymes, biological control and commercial applications. Taylor and Francis, London. 1998;185-204.
- Björkman T, Blanchard LM, Harman GE. Growth enhancement of shrunken-2 (sh2) sweet corn by Trichoderma harzianum 1295-22: effect of environmental stress. J Am Soc Hortic Sci. 1998;123(1):35-40.
- Harman GE. Myths and dogmas of biocontrol changes in perceptions derived from research on Trichoderma harzinum T22. Plant Dis. 2000;84(4):37793.
[Crossref] [Google Scholar] [PubMed]
- Ahmad JS, Baker R. Implications of rhizosphere competence of Trichoderma harzianum. Can J Microbiol. 1988;34(3):229-234.
- AppliedBiosystems. DNA Sequencing Analysis Software 6 User Guide PN 4474329A. 2014.
- Coley-Smith JR, Cooke RC. Survival and germination of fungal sclerotia. Annu Rev Phytopathol. 1971;9(1):65-92.
- Sakina A, Rather ZA, Mir S, Zargar SM, Murtaza I, Nazir N, et al. Evaluation and Validation of Rice (Oryza sativa L.) Genotypes for Tissue Culture Response from Mature Embryos. Int J Curr Microbiol App Sci. 2019;8(12):2647-2653.
- Howell CR. Mechanisms employed by Trichoderma species in the biological control of plant diseases: the history and evolution of current concepts. Plant dis. 2003 ;87(1):4-10.
[Crossref] [Google Scholar] [PubMed]
- Liu C, Shi L, Xu X, Li H, Xing H, Liang D, et al. DNA barcode goes two-dimensions: DNA QR code web server. PloS one. 2012;7(5):e35146.
[Crossref] [Google Scholar] [PubMed]
- Torelli E, Manzano M, Marks RS. Chemiluminescent optical fibre genosensor for porcine meat detection. Sens Actuators B: Chem. 2017;247:868-874.
- Romero-Arenas O, Huerta Lara M, Damián Huato MA, Domínguez Hernández F, Arellano Victoria DA. Características de Trichoderma harzianum, como agente limitante en el cultivo de hongos comestibles. Rev col de Biotecnol. 2009;11(2):143-151.
- Samuels GJ, Petrini O, Manguin S. Morphological and macromolecular characterization of Hypocrea schweinitzii and its Trichoderma anamorph. Mycol. 1994;86(3):421-435.
- Sundaramoorthy S, Balabaskar P. Biocontrol efficacy of Trichoderma spp. against wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici. J Appl Biol Biotech. 2013;1(3):036-040.
- Daniel R, Santos C, Phillips C, Fondevila M, Van Oorschot RA, Carracedo A, et al. A SNaPshot of next generation sequencing for forensic SNP analysis. Forensic Sci Int Genet. 2015;14:50-60.
[Crossref] [Google Scholar] [PubMed]
- Homma C, Inokuchi D, Nakamura Y, Uy WH, Ohnishi K, Yamaguchi H, et al. Effectiveness of blocking primers and a peptide nucleic acid (PNA) clamp for 18S metabarcoding dietary analysis of herbivorous fish. PLoS One. 2022;17(4):e0266268.
[Crossref] [Google Scholar] [PubMed]
- Gutierrez R, Roman M, Houston R, Kalafut T. Detection and analysis of DNA mixtures with the MiSeq FGx®. Sci Jus. 202;62(5):547-555.
[Crossref] [Google Scholar] [PubMed]
- Umesha S, Manukumar HM, Raghava S. A rapid method for isolation of genomic DNA from food-borne fungal pathogens. 3 Biotech. 2016;6:1-9.
[Crossref] [Google Scholar] [PubMed]
- Zhou H, Ma SJ, Song JY, Lin YL, Wu ZJ, Han ZZ, et al. QR code labeling system for Xueteng-related herbs based on DNA barcode. Chin herb med. 2019;11(1):52-59.
Citation: Baca S, Jiménez OR, González D, Huete-Pérez JA, Trabanino R, Avellaneda MC (2023) Genetic polymorphisms of seven Trichoderma spp strains, their potential as biological control agent and growth promoter in tomato. J Plant Pathol Microbiol. 14:663.
Copyright: © 2023 Baca S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

