Indexed In
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Scimago
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Review Article - (2024) Volume 15, Issue 1
Genetic Diversity and Population Structure of African Catfish (Clarias gariepinus) Species: Implications for Selection and Sustainable Genetic Improvement-A Review
Negesse Kebtieneh1,2*, Kefyalew Alemayehu3 and Gashaw Tilahun12Department of Biology, College of Natural and Computational Science, Debre Tabor University, Debre Tabor, Ethiopia
3Department of Animal Sciences, College of Agriculture and Environmental Studies, Bahir Dar University, Bahir Dar, Ethiopia
Received: 11-Dec-2023, Manuscript No. JARD-23-24296; Editor assigned: 13-Dec-2023, Pre QC No. JARD-23-24296 (PQ); Reviewed: 27-Dec-2023, QC No. JARD-23-24296; Revised: 03-Jan-2024, Manuscript No. JARD-23-24296 (R); Published: 10-Jan-2024, DOI: 10.35248/2155-9546.24.14.828
Abstract
The African catfish, or C. gariepinus, is the second-most widely cultured fish species in Sub-Saharan and most Asian nations. In order to interpret, comprehend, and manage populations and individuals, genetic diversity and population structure, as well as their quantification, are important. Due to its rapid growth rate, capacity to adapt to a variety of culture conditions, and high fertility, African catfish, C. gariepinus was first genetically improved in the 1950s and then adopted as the best catfish for African aquaculture in the middle of the 1970s. In African catfish genetics and breeding studies, a variety of molecular markers, including allozyme marker, mtDNA marker, SNPs marker, RAPD marker, Microsatellite marker and SDS-PAGE marker have been used to assess genetic similarity and divergences for insuring genetic improvement and selective breeding program of C. garipinus fish species. Genetic diversity assessment and population structure assessment are also used to quantify the genetic differentiations within and among populations of C. gariepinus fish species. These are important to formulate genetic conservation and management strategies, to sustainably manage economically important aquaculture fish species like C. gariepinus. Genetic improvement and marker assisted selective breeding programs are essential to have extensive knowledge of economically significant strains.
Keywords
African catfish, genetic diversity, population structure, genetic improvement and molecular marker
Introduction
Global aquaculture production in 2020 reached a record 122.6 million tonnes, including 87.5 million tonnes of aquatic animals worth USD 264.8 billion and 35.1 million tonnes of algae worth USD 16.5 billion. Around 54.4 million tonnes were farmed in inland waters and 68.1 million tonnes came from marine and coastal aquaculture [1]. African catfish, C. gariepinus is one of a vital inland water fish species, use for aquaculture production in parts of sub-Saharan Africa, North Africa, South America, Asia, and Europe [2]. By volume, it is the second-most widely grown fish production in Africa [1]. Because of its high fertility rate, fast growth rate even at high stocking densities, eat a variety of foods, are disease-resistant, and can withstand a variety of environmental conditions [2]. These all natural behaviors have helped African catfish, C. gariepinus spread to many areas outside of its native range particularly Thailand, Malaysia, and the Netherlands, where it was introduced for aquaculture practices and new strains like the Dutch strain were created [3-5]. African catfish, Clarias gariepinus is the most important and suitable catfish species for tropical aquaculture in Africa [5,6]. It is also generally considered to be one of the economically important freshwater fish species for rearing, whose aquaculture potential has been documented [7].
Genetic variation is beneficial and important for the long-term survival of natural populations as it ensures the provision of high fitness levels, allowing populations to adjust to new environmental conditions, and it has resulted in a phenomenon that is anticipated to be the consequence of mutation or migration to a genetically dissimilar population [8,9]. Genetic degradation is caused by a lack of understanding in the field of fish farming [10]. It happens as a consequence of inbreeding, negative selection and hybridization, resulting in an excess of homozygosity in the population and a decline in productivity. Genetic diversity is necessary for the natural population of African catfish, C. gariepinus populations to function as intended by evolution and to be preserved for future generation [8]. However, migration, genetic drift, extinction of genetically distinct wild populations and spontaneous mutation contribute to genetic changes in African catfish, C. gariepinus populations [11].
Species identification and its genetic structure are a essential issue for the economic importance of C. gariepinus. The traditional method of the characterization of Clarias gariepinus has been used to evaluate this species, and it has not been found to be a reliable method for species identification [12]. However, using molecular markers are reliable method for the genetic identification of fish species. Various authors have seen different molecular markers for determining polymorphisms based on allele frequency and amplification of DNA segments with a single primer of the arbitrary nucleotide sequence. The levels of DNA polymorphisms can be detected using molecular markers method by the presence or absence of amplification products when two strains or individuals are compared [13-17].
Several studies have been carried out to determine the genetic diversities and characterization of African catfish, Clarias gariepinus species using molecular markers such as RAPD; microsatellite markers; SNP; Isozymes; and Mitochondrial DNA marker [6,9,17-20,21-28]. However, compressive report on the genetic diversity and population structure of African catfish, Clarias gariepenus, which demonstrates for selection and sustainable genetic improvement for prospects in the future. Therefore, this review shows the report on the genetic diversity, and population structure of African catfish, Clarias gariepenus implementing for selection and sustainable genetic improvement.
Literature Review
Objective
This review objective is to assess the genetic diversity and population structure of the African catfish, C. gariepinus, for the implication of selection and sustainable genetic improvement.
Genetic diversity and population structure of African catfish
Genetic diversity: The genetic monitoring of natural populations is fundamentally important for conservation, management of natural resources, and genetic improvement programmes. Therefore, it is necessary to assess the amount of genetic diversity and the structure of diversity in samples and populations [29]. From individual fitness to ecosystem function, genetic diversity is for ecological and evolutionary processes [30]. The loss of genetic diversity in the cultured population may be due to strict breeding practices that may have genetically isolated the stock from other populations, while the loss of genetic diversity in the wild population may be due to overfishing, poaching, population division, genetic drift, and natural selection [9,18].
In order to comprehend the genetic diversity of the fish population, especially in different catfish species, it is necessary to know mean number of alleles per locus (A), mean effective number of alleles (Ae), nucleotide diversity (p), haplotype diversity (h), percentage of polymorphic loci, observed heterozygosity (Ho), expected heterozygosity (He), allelic richness (Ar), inbreeding coefficient(Fis), Shannon’s information index (Si), Unbiased expected heterozygosity (UHe), and Nei’s genetic identity (Ni) [6,8,9,11,17,19,23,27,31-33]. Genetic variation within Clarias gariepinus populations was has been reported as moderate and accounted mean number of alleles (A), mean effective number of alleles (Ae), allelic richness(Ar), observed hetrozygosity(Ho), and expected hetrozaygosity (He) ranges from 4.67-12.17; 3.15–5.80; 4.67-9.65; 0.50–0.69; 0.67-0.80, respectively [23]. Low allelic diversity was found in North African catfish populations from Thailand (A ranged from 6.00-7.00; Ae ranged from 3.43-4.59) while heterozygosity was moderate (Ho ranged from 0.52-0.72; He ranged from 0.67-0.77) [17]. Aliyu M et al, reported the effective number of alleles with in C. gariepinus populations was 1.689 from Nigeria, which is fewer than that of the reported value ranging from 0.450 ± 0.050 and 0.442 ± 0.127 between Clarias gariepinus and Heterobranchus bidorsalis populations respectively in the same country Nigeria [9,31,20].
High genetic diversity was reported within and among the endangered catfish populations in Bangladesh, and reported as the mean number of alleles (A), and the effective number of alleles (Ae), observed heterozygosity (Ho), Shannon’s information index (Si), and polymorphic information content, ranged from 41-44; 9.96-37.46; 0.57-0.76; 2.09-2.30, and, 0.84-0.88, respectively. Whereas low allelic diversity and moderate heterozygosity were reported from North African catfish, Clarias gariepinus populations in Thailand, as the mean number of alleles (A), the effective number of alleles (Ae), observed heterozygosity (Ho), and expected heterozygosity (He) ranged from 6.00-7.00; 3.43-4.59; 0.52-0.72; 0.67-0.77), respectively [17]. Similar results was reported within African catfish, Clarias gariepinus populations in Hungary as the observed and expected overall heterozygosity were between 0.519 and 0.544, respectively [22].
The mean number of alleles per locus (Na) was higher in farmed than natural C. gariepinus populations, ranging from 9.25 ± 2.9575; 6.9925 ± 2.5875 respectively, while the mean observed Heterozygosity (HO) was reported as moderate and ranging from 0.7975 ± 0.05 in natural to 0.6975 ± 0.045 in cultured C. gariepinus populations in Kenya. However, the mean expected heterozygosity ranging from 0.7675 ± 0.0475 in natural and 0.8175 ± 0.04 in cultured C. gariepinus populations were slightly higher than Ho values [25]. The genetic diversity values within farmed and wild African catfish, C. gariepinus populations were 0.4522 and 0.4018 respectively in Nigeria [9]. Whereas, the genetic diversities of bighead catfish populations were reported across three countries of Cambodia, Vietnam and Malaysia. Hence, the result showed that the higher genetic diversity in the mainland populations (Cambodia, He=0.761 to 0.813 and Vietnam, He=0.759 to 0.789 with mean number of alleles of 10.13 to 13.88) compared to Peninsular Malaysia (He=0.519 to 0.699 with mean number of alleles of 4.5 to 10.75) [34].
Moderate genetic diversity was determined in yellow catfish populations, with the observed heterozygosity (Ho) ranging from 0.42 to 0.49 and the expected heterozygosity (He) ranging from 0.51 to 0.61 in Chain [35]. The average observed heterozygosity (Ho) and expected heterozygosity (He) in the eight Channel catfish populations ranged from 0.504 to 0.767 and from 0.6 to 0.781, respectively [33]. The results showed that the eight channale catfish populations possess considerable genetic diversity.
The genetic diversity of Fare east catfish (Silurus asotus) was studied and reported as the average expected heterozygosity (He) of the wild and cultured catfish sampled populations showed were 0.907 and 0.875 respectively in Korea [32]. Similarly in Kenya, higher observed and expected heterozygosity were reported in samples of natural than the cultured African catfish, C. gariepinus populations [6]. These studies confirmed that the genetic diversity of wild or natural and cultured catfish populations was maintained at a high level. In the case of the wild catfish populations, it was effective in maintaining diversity due to the continuous fry release by the local fish research institutes. However, the genetic diversity of cultured catfish declined. Low diversity is associated with slow growth and weakened immunity. Maintaining high genetic diversity allows species to adapt to future environmental changes and avoid inbreeding, while maintain low genetic diversity the species has a small gene pool, and presences of inbreeding, which happens when there are small, isolated populations, can reduce a species’ ability to survive and reproduce [36].
Haplotype diversity and nucleotide diversities are necessary to comprehend the knowledge of genetic diversities in fish species such as catfish populations [29]. Several researchers showed each diversities within and among catfish populations such as in Indian catfish, the recorded haplotype and nucleotide diversities were found ranged from 0.06897 to 0.76322 and 0.00019 to 0.00208, respectively in India; in Clarias macrocephalu, the recorded over all haplotype (0.479) and nucleotide (0.00058) diversities were found in Philippines, which shows alarmingly low genetic diversities compared with other fresh water fish populations (0.011), in African catfish, C. gariepinus populations the nucleotide diversity was found 0.000869 in river and a tributary in northeast Nigeria, in North African catfish population, greater haplotype diversity (0.99930) and nucleotide diversity (0.07270) ware found in the three geographical isolated Rivers of Nigeria [10,26,37-39]. These genetic diversity provided important information for the development of conservation and management strategies for wild fish populations and have implications for selection and sustainable genetic improvement of fish species. Similarly, in Kenya, with in African catfish, Clarias gariepinus population the haplotype diversity was found highest in Lake Victoria (LV), and lowest in River Sosiani (SR), while, nucleotide diversity was highest in Lake Kamnarok (LKA) and lowest in Lake Victoria (LV) [40]. These show the existence of genetically distinct populations of C. gariepinus that require spatially explicit management actions such as reducing fishing pressure, pollution, minimizing habitat destruction and fragmentation for sustainable utilization of stocks.
Percentage of polymorphism, allelic richness, inbreeding coefficient, Shannon’s information index, unbiased expected heterozygosity and Nie’s genetic identity are important parameters to understand the genetic diversity of fish species [29]. The research was conducted to examine the genetic diversity of African catfish, C. gariepinus population in northeast Nigeria and reported the percentage of polymorphism from farmed and wild populations ranged from 47.3% to 75.9%, respectively and the mean numbers of inbreeding coefficient (FIS) were 0.083 and 0.053 in the farmed and wild populations, respectively [10]. The results indicating the significant level of genetic diversity in northeast Nigeria, which is useful tool for the genetic and breeding program of C. gariepinus for selection and sustainable genetic improvement.
The percentage of polymorphic loci, gene diversities and Shannon’s information index values in the three slender walking catfish, Clarias nieuhofii populations were found 75.0%, 0.2252%, and 0.3443% for SuratThani; 86.59%, 0.2982%, and 0.4441% for Narathiwat, and 96.25%, 0.3371%, and 0.5049% for Phatthalung, respectively in southern Thailand [41]. Among the 3 populations, the highest genetic distance (0.2213) was found between the Narathiwat and SuratThani populations. The results indicated a high level of genetic variation among Clarias nieuhofii from different populations in southern Thailand. This information would be useful to construct appropriate breeding programs, and could help conserve populations used as potential sources for stock management, restocking programs, and sustainable uses. Lal, Singh found that the percent polymorphism 50% and 31.25% respectively in samples from India and Thailand of exotic African catfish, Clarias gariepinus populations [27]. The results indicate that C. gariepinus in India could be the mix of different stocks belonging to different genetic lineages.
Population structure
In cases when evolutionary processes lead to genetic differentiation or genetic population structure, FST comparison is utilized as a metric by means of differentiation within and between populations as a key insight [42]. According to Holsinger and Weir, FST magnitude values range from 0 to 0.05 for minimal differentiation, 0.05-0.25 for moderate differentiation, and more than 0.25 for high population differentiation [42]. The estimated coefficients of genetic differentiations (Fst) and gene flow (Nm) values are used to estimate the differences between populations and determine the genetic structure of catfish. Hence, different authors reported the FST and Nm values in different catfish populations, (FST=0.1622, Nm=1.2909 [19] for African catfish, Calarias gariepinus; (FST=0.130, Nm=0.298-47.786) [34], for Bighead catfish Clarias macrocephalus; (FST=0.038, Nm=12.79 [16]) for Bighead catfish; (FST=0.2815, Nm=1.2762 [41], for Slender Walking Catfish, Clarias nieuhofii; (FST=ranging from 0.203-0.129, Nm=0.9490) [9], for African catfish, Clarias gariepinus; (FST=0.011, Nm=24.333) (Table 1) [16,21,34,41]. These indicate that there are significant factors between populations and gene flows are the main genetic differentiations.
| Country | Species | Sources of variations in% | Estimated mean FST values | Estimated genetic flow (Nm) | Estimated genetic distances (D) | Over all differentiation | References | |
|---|---|---|---|---|---|---|---|---|
| Among populations | Within populations | |||||||
| Kenya | African catfish, Clarias gariepinus | 22 | 88 | 0.22 | - | - | Moderate | [25] |
| Nigeria | African catfish , Clarias gariepinus | 1 | 99 | 0.19 | - | 0.5213 | Moderate | [46] |
| India | Indian catfish, Clarias magur | 34.01 | 65.99 | 0.34014 | - | - | High | [37] |
| Thailand | African catfish, Calarias griepinus | - | - | 0.144 ± 0.026 | - | 0.036 to 0.144 | Moderate | [17] |
| Thailand | North African catfish, Clarias gariepinus | 15.76 | 84.24 | 0.096 ± 0.034 | - | 0.087 to 0.161 | Moderate | [17] |
| India | African catfish Clarias gariepinus | 18 | 82 | 0.1622 | 1.2909 | 1.5798 to 3.4526 | Moderate | [19] |
| Kenya | African catfish, Clarias gariepinus | 17.22 | 86.69 | 0.133 | - | - | Moderate | [40] |
| Malasia | Bighead catfish Clarias macrocephalus |
13.37 | 86.63 | 0.130 | 0.298 to 47.786 | - | High | [34] |
| Nigeria | African catfish, Clarias gariepinus | 8 | 92 | 0.01 | - | - | Low | [13] |
| Viet Nam | Bighead catfish | - | - | 0.038 | 12.79 | 0.007 to 0.019 | Low | [16] |
| Thailand | Slender Walking Catfish, Clarias nieuhofii | - | - | 0.2815 | 1.2762 | 0.1381 to 0.2213 | High | [41] |
| Nigeria | North African catfish, Clarias gariepinus | 75.73 | 24.27 | 0.75726 | 5.850 | 0.008 to 0.0519 | high | [10] |
| Nigeria | African catfish Clarias gariepinus | 72 | 28 | 0.719 | 0.098 | - | high | [13] |
| Bangladesh | Endangered catfish, Rita rita | 1 | 99 | 0.11 | 24. 333 | 0.120 to 0.256 | low | [21] |
| Nigeria | African catfish Clarias gariepinus | 4 | 96 | 0.203 to 0.129 | 0.9490 | 0.1038 | Variable | [8] |
| Philippines | Clarias macrocephalus | 80.05 | 19.95 | 0.80050 | - | - | high | [38] |
| Thailand | Striped catfish, Pangasianodon hypophthalmus | 17.57 | 82.4 | 0.167 | - | 0.055 to 0.148 | moderate | [45] |
| China | Yellow catfish, Pelteobagrus fulvidraco | 3.10 | 96.90 | 0.031 | - | 0.080 to 0.273 | Variable | [44] |
Table 1: Estimated sources of genetic variations, coefficients of genetic differentiations and average gene flow of different catfish populations.
Genetic distance is a measure of the genetic divergence between species or between populations within a species, whether the distance measures time from common ancestor or degree of differentiation [43]. Hence, genetic distance (D) also showed the significant genetic differentiation within and among groups of catfish populations and determines the population structure. Genetic distance between populations of catfish reported by different authors reviewed as showed in Table 1. Genetic distances varying from 0.036-0.144 between population in the species of African catfish, Clarias gariepinus in Thailand; varying from 0.087-0.161 between populations in the same species in the other part of the same country; from 1.5798-3.4526 between populations in African catfish in India, from 0.007-0.019 between populations of bighead catfish in Viet Nam; from 0.1381-0.2213 between populations of Slender Walking Catfish, Clarias nieuhofii in Thailand; from 0.008-0.0519 between population of North African catfish, Clarias gariepinus in Nigeria, from 0.120-0.256 between populations of Endangered catfish, Rita rita in Bangladesh, from 0.080-0.273 between populations of Yellow catfish, Pelteobagrus fulvidraco in north to south Chain; from 0.055-0.148 between populations of striped catfish, Pangasianodon hypophthalmusin Thailand; 0.5213 between populations of African catfish , Clarias gariepinusin Nigeria; 0.1038 between populations of African catfish, Clarias gariepinus in Nigeria [9,10,16,17,21,44-46]. These indicate that catfish populations with many similar alleles have small genetic distances. While catfish populations with different allele may have high genetic distances.
The applications of molecular markers for genetic diversity and population structure of African catfish: The most commonly used molecular markers in aquaculture include Allozymes, mitochondrial DNA (mtDNA), Restriction Fragment Length Polymorphism (RFLP), Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphism (AFLP), microsatellites, Expressed Sequence Tag (EST) and Single Nucleotide Polymorphism (SNP) [47]. Microsatellite marker, RAPD, SNPs, mitochondrial DNA (mtDNA), and Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDSPAGE) are used for African catfish, C. gariepinus species genomic studies (Table 2).
| Species | Molecular markers | Applications | References |
|---|---|---|---|
| North African Catfish, Clarias gariepinus | mtDNA (mitochondrial DNA) | To study the population genetic structure and genetic diversity of wild and farmed African catfish, Clarias gariepinus fish species populations in Uganda | [48] |
| African Catfish, Clarias gariepinus |
SNPs (Single Nucleotide Polymorphism) | To distinguish Clarias gariepinus from its closest relative Clarias anguillaris in Nigeria | [24] |
| North African Catfish, Clarias gariepinus | mtDNA (mitochondrial DNA) | To demonstrated the population genetic differentiation and genetic diversity of C. gariepinus in the three water bodies of Southwest Nigeria, which provided information for the development of conservation and management strategies for wild fish populations. | [10] |
| African Catfish, Clarias gariepinus |
RAPD (Random Amplified Polymorphic DNA) | To assess the population genetic structure, genetic diversity and allelic richness of wild and farmed African catfish populations in Nigeria. | [8] |
| African Catfish, Clarias gariepinus |
RAPD (Random Amplified Polymorphic DNA) | To characterize the genes of wild and farmed African Catfish populations, useful for genetic and breeding program of Clarias gariepinus in northeastern Nigeria. | [8] |
| Exotic African catfish, Clarias gariepinus | Allozyme | To distinguish the genetic diversity of exotic catfish, Clarias gariepinus sampled from in India and Thailand | [27] |
| African Catfish, Clarias gariepinus |
mtDNA (mitochondrial DNA) and Microsatellite | To demonstrate the genetic diversity and population structure of C. gariepinus population, which used to conservation and aquaculture practices in Kenya. | [27] |
| African Catfish, Clarias gariepinus | SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) |
To assess the genetic diversity of African catfish population from the two lakes in Nigeria. | [9] |
| African Catfish, Clarias gariepinus | Microsatellite | To demonstrate the genetic variations and differentiations, useful for analysis of genetic diversity and management of the fish resources in India. | [19] |
| African Catfish, Clarias gariepinus | Microsatellite | For strain selection in Thailand | [23] |
| African Catfsh, Clarias gariepinus | Microsatellite | To analyze the genetic diversity and population structure of African catfish populations, useful for population and conservation genetics of natural and bred African catfish populations in Hungary. | [22] |
| North African catfish, Clarias gariepinus | Microsatellite | To analysis the genetic diversity and population structure of North African Catfish, which are useful for establishing a base population for a genetic improvement program in Thailand. | [23] |
| Clarias gariepinus (Siluriformes, Clariidae) | Microsatellite | For separation of populations, this used to the analysis of genetic diversity and population structure of C. gariepinus populations in Nigeria. | [13] |
| African catfish, Clarias gariepinus |
RAPD (Random Amplified Polymorphic DNA) and Microsatellite | To analysis the genetic diversity of wild and cultured African catfish populations in Nigeria. | [13] |
Table 2: Recently published applications of molecular markers in genetic diversity and population structure of African catfish, C. gariepinus species.
SNPs are the most widely used marker in African Catfish C. gariepinus fish population genetics study since they are found in abundance. Isa SI et al, used SNPs marker to identify Clarias gariepinus from its closest relative Clarias anguillaris from Nigeria, for conservation and management purposes [24]. Mitochondrial DNA marker is used for study the population genetic structure and genetic diversity of wild and farmed African catfish, Clarias gariepinus fish species populations in Uganda; for analysis the population genetic differentiation and genetic diversity of C. gariepinus in the three water bodies of Southwest Nigeria and for analysis the genetic diversity and population structure of C. gariepinus population in Kenya [6,10,24,48]. Random Amplified Polymorphic DNA (RAPD) is also used for the genomics studies of African catfish, Clarias gariepinus species for different purposes like for the assess the population genetic structure, genetic diversity and allelic richness of wild and farmed African catfish populations in Nigeria; for gene characterization of wild and farmed African Catfish populations, C. gariepinus, which useful for genetic and breeding program in northeastern Nigeria and for analysis of genetic diversity of wild and cultured African catfish, Clarias gariepinus populations in Nigeria [8,9,18]. Aliyu M et al, showed Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS PAGE) marker used to assess the genetic diversity of African catfish, C. gariepinus populations from the two lakes in Nigeria [31].
Microsatellite markers are used increasingly in aquaculture fish species due to their elevated Polymorphic Information Content (PIC), co-dominant mode of expression, mendelian inheritance, abundance and broad distribution throughout the genome [49]. Microsatellite markers have also been developed in Claris gariepinus fish species for desirable traits like growth enhancement, disease resistance, etc. [24]. Some authors used microsatellite marker for different genomic studies in African catfish, Calarias gariepinus species like demonstrating genetic differentiation and population structure in Kenya; for genetic variations and differentiations in India; for strain selection in Thailand; for analysis the genetic diversity and population structure in Hungary; for analysis the genetic diversity and population structure of North African catfish in Thailand, for separation of Clarias gariepinus (Siluriformes, Clariidae) fish populations and for analysis the genetic diversity of wild and cultured populations in Nigeria as indicated in Table 2 [6,17-19,22,23,46]. Microsatellites have high Polymorphism Information Content (PIC) values for which they are considered very reliable. These molecular markers have application while comparing the wild and hatchery fish population. Hence for a sustainable management and genetic improvement of Claria gariepinus fish resources it is imperative to utilize molecular markers.
Trial I: Marker assisted selective breeding in African catfish, C. gariepinus fish species
Marker-Assisted Selection (MAS) is the process of using morphological, biochemical, or DNA markers as indirect selection criteria for selecting important traits in fish breeding [50]. This process is used to improve the effectiveness or efficiency of selection for the traits of interest in breeding programs.
The choice of economically important traits for genetic improvement largely depends on the consumers preference, the farmers (yield, growth, diseases resistances and survival) and the feasibility of techniques or methods involved in farmed fish species like Clarias gariepinus species [51]. For instances, in Indonesia, the mutiara strain of African catfish, Clarias gariepinus was performed 10%-40% better in growth, 15%-70% better in productivity, 2-9 times higher in benefit-cost ratio, shorter growing period (45-60 days), lower feed conversion ratio (0.6-0.8) in nursery and (0.6-1.0) in grow out) and higher survival rate (60%-70%) and it was much more resistant to disease than the local strain counterpart, useful for genetic improvement program [52].
Selective breeding for increased production is expected to enhance efficiency of resource utilization of a production system, through correlated changes in feed efficiency or shorter production period [53,54]. Common steps using a selective breeding program of aquaculture fish species including African catfish, C. gariepinus are presented by Figure 1.
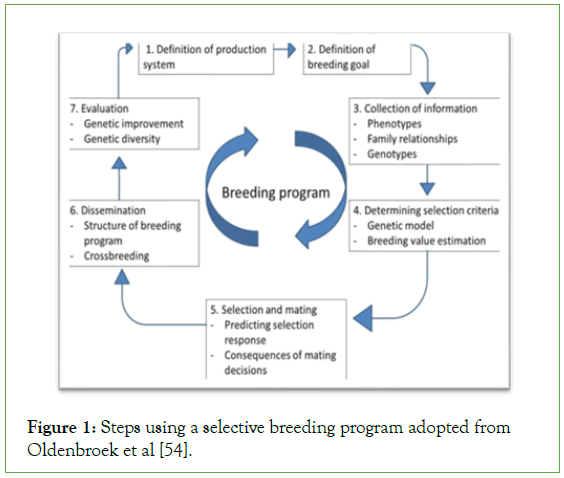
Figure 1: Steps using a selective breeding program adopted from Oldenbroek et al [54].
Sustainable genetic improvement: According to estimates, just 10% of the world’s aquaculture industry uses genetically modified stocks [55]. Choice of breeds, cross breeding, and selective breeding are some of the current techniques for genetic improvement used in aquaculture, while sex reversal, sterilization, and triploidization are more recent techniques used to improve fish production in aquaculture industry [52]. Genome-editing technologies such as CRISPR/Cas9 experiments with cultured fishes have focused on improving growth rate and disease resistance, achievement of reproductive confinement, and other valued traits [56,57].
Infectious disease is one of the primary constraints to aquaculture production and, therefore, a major target for selective breeding and genome-editing approaches [57]. Host resistance to certain pathogens is a suitable trait for the use of genome-editing technologies due to the difficulty in non-destructive measurement of the trait in breeding candidates, the plausibility of utilizing cell culture genome-wide pooled CRISPR screens, and the frequent availability of early life in vivo-established challenge models [58].
Different categories of genome-editing applications include: (i) discovery of causative variants underlying single or multiple Quantitative Trait Loci (QTLs) affecting traits of interest, and subsequent fixation of the favorable alleles using editing; (ii) introgression-by-editing of favorable alleles into closed breeding systems from other populations, strains, or species; and (iii) creation and use of de novo alleles with positive effects on the trait of interest.
The Genome-editing technologies, such as CRISPR/Cas9, would be useful for the industry to assemble the genomes of strains of Clarias macrocephalus and Clarias gariepinus cultured in Thailand to the chromosome level and annotate the protein pathways relevant to aquacultures, such as disease resistance, growth, development, adaptation, and sex determination, by developing a number of marker candidates. Therefore, Clariid bio resources are important for genetic improvements in sustainable catfish farming [59]. The other report revealed that gene editing of channel catfish for the reproductive confinement of gene-engineered, domestic, and invasive fish to prevent gene flow into the natural environment appears potential [60]. These are important for genetic improvement of Clarias gariepinus provides potential in improving production. Strains of African catfish, C. gariepinus species that grow faster, exhibit greater disease resistance, and have other more favorable characteristics for aquaculture have been produced through selective breeding [52].
Economically significant genetic strains of Clarias gariepinus species include those that exhibit traits associated with aquaculture performance, such as growth, Feed Conversion Ratio (FCR), productivity, resistance to disease, resistance to stress, size uniformity, and benefit/cost ratio, used for the sustainable genetic improvement for the future generations [52]. Catfish production qualities have been successfully improved using selection, intraspecific crossbreeding, interspecific hybridization, and genetic engineering [61]. In contrast to this, other breeding programs, interspecific hybridization actually enhanced more characteristics in a single cross. However, more genetic improvement was produced by mass selection combined with crossbreeding, mass selection combined with genetic engineering, or mass selection combined with strain selection and hybridization than by any other genetic improvement program [62].
Knowledge gaps and challenges
Much progress has been made on the studies of genetic diversity and population structure of C. gariepinus species, which are useful for sustainable genetic improvement, there are still knowledge gaps and challenges that need to be addressed.
Conclusion and Recommendations
From this review we have investigated, African catfish, Claris garirpinus fish genetic diversity and population structure assessments done by using molecular markers such as Microsatellites, SNPs, allozyme analysis, RAPD, mitochondrial DNA and SDS-PAGE markers as a useful tool to formulating meaningful conservation, and management strategies of commercially important fish species, which is African catfish. The study of genetic diversity and population structure is also important tool for selective breeding program, which used to sustainable genetic improvement for the next generations by providing economically important genetic strains like rapid increasing growth rate, disease resistance traits, increasing meat quality and etc. understanding the genetic diversity and population structure have a sinificant role to maintain a diverse and representative brood stock populations from various sources and geographical areas to preserve genetic diversity. This indicates that regular monitoring and managing brood stock populations to minimize inbreeding depression and maintain genetic diversity. Employ advanced breeding strategies, including mate selection, genome-wide selection, and marker-assisted selection, to accelerate genetic improvement while maintaining genetic diversity.
Develop strategies for the conservation of genetically unique and endangered African catfish populations. Consider controlled hybridization between different populations, if appropriate, while ensuring genetic integrity and avoiding negative impacts on wild populations. Establish a system for continuous genetic monitoring of hatcheries, farms, and wild stocks to ensure the preservation and improvement of genetic diversity. Regularly report these findings to relevant stakeholders, including researchers, breeders, and policymakers. Foster collaboration between researchers, breeders, and stakeholders to share knowledge, exchange genetic resources, and implement sustainable breeding practices. Invest in capacity building programs to train professionals involved in genetic improvement activities.
Acknowledgement
The authors would like to extend their gratitude to Bahir Dar and Debre Tabor universities, giving the chance to review this paper.
Conflicts of Interest
There are no conflicts of interest.
References
- Fisheries FA. The state of world fisheries and aquaculture. Towards blue transformation.2022.
- Thirukanthan CS, Azra MN, Piah RM, Lananan F, Téllez-Isaías G, Gao H, et al. Catfishes: A global review of the literature. Heliyon. 2023;9(9):e20081
[Crossref] [Google Scholar] [PubMed]
- Nwafili SA, Gao T. Is the Dutch domesticated strain of Clarias gariepinus (Burchell, 1822) a hybrid? Afr J Biotechnol. 2007;6(8).
- Hanssens M. A review of the Clarias species (Pisces; Siluriformes) from the lower Congo and the pool Malebo. 2009.
- Ouma DF, Barasa JE. Perspective chapter: Species diversity and distribution of catfishes and their current contribution to global food security. InCatfish-Advances, Technology, Experiments 2022. IntechOpen.
- Barasa JE, Mdyogolo S, Abila R, Grobler JP, Skilton RA, Bindeman H, et al. Genetic diversity and population structure of the African catfish, Clarias gariepinus (Burchell, 1822) in Kenya: Implication for conservation and aquaculture. 2017;147(2).
- Dauda AB, Natrah I, Karim M, Kamarudin MS, Bichi AU. African catfish aquaculture in Malaysia and Nigeria: Status, trends and prospects. Fish aquac j. 2018;9(1):1-5.
- Suleiman IO, Moruf RO, Usman BI. Population genetic structure of feral and cultured African Catfish (Clarias gariepinus) inferred from random amplified polymorphic DNA in Kano, Nigeria. Trop J Nat Prod Res. 2023;7(3).
- Suleiman SB, Diyaware MY, Aliyu M, Mohammed ZB. Genetic characterization of farmed and wild populations of African catfish (Clarias gariepinus Burchell, 1822) using the random amplified polymorphic marker. J Agric Sci, Belgrade. 2020;65(4):375-389.
- Popoola OM. Genetic differentiation and molecular phylogenetics of North African Catfish from three distinct waterbodies. Croatian Journal of Fisheries. 2022;80(3):123-132.
- Bassey H. Phenotypic and genetic evaluation of farmed and wild Clarias gariepinus broodstocks in Nigeria. 2020.
- Ikpeme EV, Udensi OU, Okolo MC, Ogban FU, Ufford NG, Odo EU, et al. Genetic relatedness of Clarias gariepinus (L.) from cultured and wild populations using multivariate analyses. Asian J Anim Sci. 2016;10(2):131-138.
- Ola-Oladimeji FA, Awodiran MO, Olaleye VF, Awopetu JI. Genetic characterization based on RAPD-PCR in cultured strains of Clarias gariepinus (Siluriformes: Clariidae). Genetics of Aquatic Organisms. 2020;4(2):81-88.
- Okumuş İ, Çiftci Y. Fish population genetics and molecular markers: II-molecular markers and their applications in fisheries and aquaculture. Turkish J Fish Aquat Sci. 2003;3(1).
- Michael PO. Genetic differentiation and molecular phylogenetics of North African catfish from three distinct waterbodies. 2022.
- Nguyen NT, Duong TY. High genetic diversity and gene flow among cultured and wild populations of bighead Catfish (Clarias macrocephalus) in the Mekong Delta of Viet Nam inferred from ISSR markers. J Fish Environ. 2022;46(2):67-76.
- Wachirachaikarn A, Na-Nakorn U. Genetic diversity of the North African catfish, Clarias gariepinus (Burchell, 1822) hatchery stocks in Thailand. Science Asia. 2019;45(4).
- Awodiran MO, Afolabi O. Genetic diversity in cultured and wild population of Clarias gariepinus (Burchell, 1822) in Nigeria using random amplified polymorphic DNA (RAPD) and microsatellite DNA. Fish Aquac J. 2018;9:247.
- Ezilrani P, Christopher JG. Genetic variation and differentiation in African catfish, Clarias gariepinus, assessed by heterologous microsatellite DNA. 2015
- Agbebi OT, Ilaboya DE, Adebambo AO. Preliminary characterization of genetic strains in Clariid species, Clarias gariepinus and Heterobranchus bidorsalis using microsatellite markers. Afr J Biotechnol. 2013;12(4):364.
- Ali MF, Salam MA, Sarder M, Rahman M, Mollah M. Genetic diversity and population structure of endangered Catfish Rita rita (Hamilton, 1822) revealed by heterologous DNA microsatellite markers. Asian Fish Sci. 2021;34(2).
- Kánainé Sipos D, Bakos K, Ősz Á, Hegyi Á, Müller T, Urbányi B, et al. Development and characterization of 49 novel microsatellite markers in the African catfish, Clarias gariepinus (Burchell, 1822). Mol Biol Rep. 2019;46:6599-6608.
- Wachirachaikarn A, Rungsin W, Srisapoome P, Na-Nakorn U. Crossing of African catfish, Clarias gariepinus (Burchell, 1822), strains based on strain selection using genetic diversity data. Aquaculture. 2009;290(1-2):53-60.
- Isa SI. Development of genetic improvement in the African Catfish (Clarias gariepinus, Burchell, 1822). 2019.
- Echessa JB. Genetic diversity, population structure and influence on life-history traits of the African Catfish, Clarias gariepinus (Burchell 1822), in Kenya (Doctoral dissertation, University of Eldoret). 2018
- Uruku MN, Adikwu IA, Oyebola OO, Akombo PM. Genetic diagnosis on strains of the African Catfish, Clarias gariepinus (Burchell 1822) in River Benue and a Tributary in North East Nigeria. 2021.
- Lal KK, Singh RK, Mohindra V, Singh B, Ponniah AG. Genetic makeup of exotic catfish Clarias gariepinus in India. Asian Fish Sci. 2003.
- Nyunja C, Maina J, Amimo J, Kibegwa F, Harper D, Jung'a J. Stock structure delineation of the African Catfish (Clarias gariepinus) in selected populations in Kenya using mitochondrial DNA (Dloop) variability. J Aquac Res. 2017;8:485.
- Hvilsom C, Segelbacher G, Ekblom R, Fischer MC, Laikre L, Leus K, et al. Selecting species and populations for monitoring of genetic diversity. IUCN Publication. 2022.
- Sjöqvist CO, Kremp A. Genetic diversity affects ecological performance and stress response of marine diatom populations. ISME J. 2016;10(11):2755-2766.
- Aliyu M, Diyaware MY. Genetic diversity of African catfish (Clarias Gariepinus Burchell, 1822) from lakes alau and Maladumba using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE). Niger J Anim Sci Technol. 2019;2(2):46-55.
- Kim JE, Hwang JA, Kim HS, Lee JH. Genetic variability comparison of wild and cultured far eastern catfish (Silurus asotus) of Korea using microsatellite marker. Dev Reprod. 2020;24(4):317.
- Ma Q, Wu B, Jiang J, Song Z. Genetic characterization of selected domestic populations of channel catfish (Ictalurus punctatus) using microsatellites. Pakistan J. Zool. 2020;52:1683-1689.
- Nazia AK, Tam BM, Jamaluddin JA, Nor SA. High genetic structure between natural populations of bighead catfish Clarias macrocephalus (Günther, 1864) from the Mekong Delta and Peninsular Malaysia. Fish Res. 2021;241:105993.
- Zhong L, Wang M, Pan J, Li D, Tang S, Bian W, et al. Evaluation of genetic diversity and population structure of five yellow catfish populations by microsatellite markers. Oceanol Hydrobiol Stud. 2018;47(2):99-106.
- Neff BD, Garner SR, Pitcher TE. Conservation and enhancement of wild fish populations: Preserving genetic quality versus genetic diversity. Can J Fish Aquat. 2011;68(6):1139-1154.
- Sahoo L, Barat A, Sahoo SK, Sahoo B, Das G, Das P, et al. Genetic diversity and population structure of endangered Indian catfish, Clarias magur as revealed by mtDNA D-loop marker. Turkish J Fis Aqua Sci. 2020;21(1):9-18.
- Tan MT, Jumawan JC, Quilang JP. Low genetic diversity in Clarias macrocephalus Günther, 1864 (Siluriformes:Clariidae) populations in the Philippines and its implications for conservation and management. J Threat Taxa. 2016;8(6):8849-8859.
- Manel S, Guerin PE, Mouillot D, Blanchet S, Velez L, Albouy C, et al. Global determinants of freshwater and marine fish genetic diversity. Nat Commun. 2020;11(1):692.
- Alal GW, Barasa JE, Chemoiwa EJ, Kaunda‐Arara B, Akoll P, Masembe C. Genetic diversity and population structure of selected lacustrine and riverine populations of African catfish, Clarias gariepinus (Burchell, 1822), in Kenya. J Appl Ichthyol. 2021;37(3):427-438.
- Pechsiri J, Vanichanon A. Genetic diversity in slender walking catfish (Clarias nieuhofii) populations: Implications for population management. Walailak J Sci & Tech. 2016;13(7):511-519.
- Holsinger KE, Weir BS. Genetics in geographically structured populations: Defining, estimating and interpreting F ST. Nat Rev Genet. 2009;10(9):639-650.
- Wei H, Geng L, Shang X, Li L, Ma B, Zhang Y, et al. Comparison genetic diversity and population structure of four Pseudaspius leptocephalus populations in Heilongjiang River Basin based on mitochondrial COI gene. Front Mar Sci. 2023;10:1158845.
- Guo XZ, Chen HM, Wang AB, Qian XQ. Population genetic structure of the yellow catfish (Pelteobagrus fulvidraco) in China inferred from microsatellite analyses: Implications for fisheries management and breeding. J World Aquac Soc. 2022;53(1):174-191.
- Na-Nakorn U, Moeikum T. Genetic diversity of domesticated stocks of striped catfish, Pangasianodon hypophthalmus (Sauvage 1878), in Thailand: Relevance to broodstock management regimes. Aquaculture. 2009;297(1-4):70-77.
- Awodiran MO, Adeniran FO, Akinwale RO, Akinwande AA. Microsatellite variability of two populations of Clarias gariepinus (siluriformes, Clariidae) in Nigeria. Vestnik Zoologii. 2019;53(3):195-208.
- Rodrigues KF, Biun H, Yong WT, Chin GJ, Ching FF, Othman R. The application of molecular markers in fish breeding and aquaculture. InMarine Biotechnology: Applications in food, drugs and energy. 2023:73-101.
- Daniel O. Population genetics structure of wild and domesticated African catfish (Clarias gariepinus) in Victoria and Albertine drainage basins in Kenya. 2015.
- Martinez V, Guimarães E, Ruane J, Scherf B, Sonnino A, Dargie J. Marker-assisted selection in fish and shellfish breeding schemes. Marker-Assisted selection-Current status and future perspectives in crops, livestock, forestry and fish. 2007:329-362.
- Haldar C. Importance of marker assisted selection in fish breeding in India: Challenges and opportunities. Ann Aquac Res. 2018;5(1):1047.
- Eze F. Marker-assisted selection in fish: A review. Asian J Fish Aquat. 2019;3(4):1-11.
- Imron I, Iswanto B, Suparapto R, Marnis H. Development of genetically improved farmed African Catfish, Clarias gariepinus; a review and lessons learned from Indonesian fish breeding program. InIOP Conference Series: Earth and Environmental Science. IOP Publishing. 2020;593(1):012032.
- Sae-Lim P, Kause A, Mulder HA, Olesen I. Selective breeding in aquaculture for future environments under climate change. Internal project View project OrAqua View project. 2016.
- Oldenbroek K, van der Waaij L. Textbook animal breeding: Animal breeding and genetics for BSc students. 2014.
- Gjedrem T, Robinson N, Rye M. The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture. 2012;350:117-129.
- Hallerman E. Genome editing in cultured fishes. CABI Agric Biosci. 2021;2(1):1-9.
- Gratacap RL, Wargelius A, Edvardsen RB, Houston RD. Potential of genome editing to improve aquaculture breeding and production. Trends Genet. 2019;35(9):672-684.
- Gaj T, Sirk SJ, Shui SL, Liu J. Genome-editing technologies: Principles and applications. Cold Spring Harb Perspect Biol. 2016;8(12):a023754.
- Lisachov A, Nguyen DH, Panthum T, Ahmad SF, Singchat W, Ponjarat J, et al. Emerging importance of bighead catfish (Clarias macrocephalus) and north African catfish (C. gariepinus) as a bio resource and their genomic perspective. Aquaculture. 2023:739585.
- Qin G, Qin Z, Lu C, Ye Z, Elaswad A, Bangs M, et al. Gene editing of the catfish gonadotropin-releasing hormone gene and hormone therapy to control the reproduction in channel catfish, Ictalurus punctatus. Biology. 2022;11(5):649.
- Houston RD, Kriaridou C, Robledo D. Animal board invited review: Widespread adoption of genetic technologies is key to sustainable expansion of global aquaculture. Animal. 2022;16(10):100642.
- Dunham RA, Liu Z. Gene mapping, isolation and genetic improvement in catfish. Aquatic genomics: Steps toward a great future. 2003:45-60.
Citation: Kebtieneh N, Alemayehu K, Tilahun G (2023) Genetic Diversity and Population Structure of African Catfish (Clarias gariepinus) Species: Implications for Selection and Sustainable Genetic Improvement. A Review. J Aquac Res Dev. 15:828.
Copyright: © 2024 Kebtieneh N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.