Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- ResearchBible
- Cosmos IF
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2019) Volume 8, Issue 1
Genetic Diversity Analysis of Walnut (Juglans regia L.) from Kashmir Valley Using RAPD and ISSR Markers
Uzma Noor Shah1, JI Mir2, N Ahmed3 and Khalid M Fazili1*2Central Institute of Temperate Horticulture (ICAR), Srinagar, J&K, India
3Sher-i-kashmir University of Agricultural Science and Technology, J&K, India
Received: 28-Jul-2019 Published: 10-Sep-2019, DOI: 10.35248/2168-9881.19.8.185
Abstract
Walnut (Juglans regia) is one of the most essential nutritive and economic nut crops. Genetic diversity analysis of 96 walnut genotypes from Kashmir was done using 13 RAPD and 19 ISSR markers. RAPD diversity analysis revealed 27.78% polymorphism while as ISSR analysis showed 93.05% of polymorphic bands. Jaccards similarity co-efficient ranged from 0.78 to 1 with an average of 0.91 for RAPD and 0.22 to 0.85 with an average of 0.58 for ISSR markers and 0.59 to 0.92 with an average of 0.74 for combined data. Cluster analysis using UPGMA by RAPD and ISSR markers obviously revealed distinct diversity between genotypes. The dendrogram constructed from the combination of RAPD and ISSR markers categorized the 96 genotypes into one major cluster and five minor clusters. Effective number of alleles (Ne), Nei’s gene diversity (H) and Shannon’s Information index (I) of 96 samples were 1.12, 0.08 and 0.12 respectively for RAPD data and 1.48, 0.29 and 0.44 for ISSR data respectively. Positive correlation (r=0.11) was observed between RAPD and ISSR markers using the Mantel test and thus supporting the validity of the dendrogram. The high number of alleles and a broad range of genetic diversity were observed among all the genotypes, accounting them as potential candidates for selective breeding for particular traits and expanding the genetic base. Based on our results, RAPD and ISSR analysis can be used for the characterization and categorizing of walnut genotypes and indicate that Jammu and Kashmir has a significant genetic diversity pool and rich genetic variance of walnuts.
Keywords
Juglans regia; RAPD markers; ISSR markers; Molecular characterization; Genetic diversity
Introduction
The genus Juglans is characterized by monoecious and heterodichogamous character, counting approximately 40 species, caterogised into three sections, Dioscaryon Dode, Cardocaryon Dode and Thyoscaryon Dode. Juglans regia L. belongs to Dioscaryon Dode Section and is the most economically important nut and oil species. Walnut grows well in regions having temperate climate and is naturally distributed from east of Turkey to north of Pakistan, Iran, Afghanistan, mountains of Nepal and Central Asia [1]. In year 2014/15, worldwide production of walnuts reached 655,651 metric tons (kernel basis), 14 percent high from the previous year, and 87 percent high when compared with 2004/05, which confirms the upward drift over the last decade [2]. The China and United States clearly led global production, accounting for the 67 percent of total world production. India contributes 2% to the world’s total walnut production. Worldwide production of walnut has been rising speedily in recent years mainly in Asian countries due to its high nutritional value and antioxidant properties [3]. In India, walnuts are grown in Arunachal Pradesh, Himachal Pradesh, Uttarakhand and Jammu and Kashmir. J&K is the main centre for commercial walnut production in India and contributes around 98% of the country’s output and the state has been declared as the “Agri. Export zone for Walnuts”. Most walnuts produced for export by India are produced in this state. According to the United Nations Development Programme Special Unit for South-South Cooperation (UNDP SU/SSC), 63,000 hectares of Jammu and Kashmir are presently under walnut cultivation. These plantings produce around 60,000 tonnes of walnuts worth an approximated 25 million rupees. Moreover, the plant has medicinal importance for human health which is because of its high antioxidant capacity [4] and ω-3 fatty acid concentration [5].
A thorough understanding of population diversity is must for the valuable utilization of the genetic variability accessible to breeders [6]. In order to raise the walnut production and exportation, new genotypes should be introduced and old genotypes should be preserved in gene banks [7]. For achieving this goal, the foremost step is detection of superior genotypes and then characterization of their genetic variety. For this, the conventional methods are now being complemented by molecular methods, enabling breeders to take better decisions while choosing the germplasm to be used in breeding programs [8]. Therefore molecular markers are suggested to explore genetic diversity in walnut [9]. Reports are available concerning evaluation of genetic diversity among walnut genotypes using Restriction Fragment Length Polymorphism (RFLP) [10], Random Amplified Polymorphic DNA (RAPD) [11], Inter- Simple Sequence Repeat (ISSR) markers [12,13], Microsatellite markers [9,14] and Amplified Fragment Length Polymorphism (AFLP) [15]. Ultimately, high-quality genotypes could be used in crossing programs to produce new commercial genotypes.
RAPD markers have the advantages of being easy to use, low cost of experiments and they also cover the entire genome. Moreover, they can be used as a primary method of choice for screening the genotypes to find duplicates in collections. RAPDs have been used to evaluate the level of polymorphism in Juglans [7] as well as in other important fruit tree species, with attractive results [16]. In Jammu and Kashmir, India, a high level of genetic diversity was found within walnut populations exhibited by Morphological, RAPD and SSR markers [17,18].
ISSRs are important tool to assess the genetic diversity in a plant collection or wild species, and are helpful to identify genotypes, even in highly related individuals [12,19-21]. In addition to the reproducibility and low cost of the technique [22], the most important advantage of this method is its universality and ease of development [6], ISSRs have been extensively used to evaluate genetic diversity of populations of economic forest tree species [23]. To our knowledge, ISSR have never been used in walnut genetic diversity studies in India. So we are reporting it for the first time. The aim of this study was to find out the genetic relationships among 96 genotypes of Juglans regia from different selected districts of Jammu and Kashmir with different molecular markers.
Material and Methods
Plant material
A total of ninety six walnut genotypes were obtained from the research farm of Central Institute of Temperate Horticulture, Srinagar, India representing 94 local selections from eight geographically isolated districts of Jammu And Kashmir State and two exotic varieties (Table 1). Maximum number (32) of genotypes were represented by district Budgam followed by Ganderbal (26), Pulwama (11), Baramulla (11), Anantnag (07), Shopian (03) Srinagar (03) and Bandipora (01).
Table 1: Details of 96 walnut genotypes used in the study.
| S. No | Accession No. | Collection Site | Latitude | Longitude | Altitude | |
|---|---|---|---|---|---|---|
| 1 | AMC-5 | IC587092 | Anantnag | 33°73 | 75°16 | 5,253 ft |
| 2 | GSS-18 | IC587139 | Ganderbal | 34°23 | 74°78 | 5,302 ft |
| 3 | UB-3 | IC587210 | Baramulla | 34°19 | 74°36 | 5,226 ft |
| 4 | LG-10 | IC561052 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 5 | APS-13 | IC587272 | Anantnag | 33°73 | 75°15 | 5,253 ft |
| 6 | GKS-1 | IC587142 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 7 | SHS-1 | IC587175 | Shopian | 33°9 | 74°99 | 5600ft |
| 8 | SPB-2 | IC587299 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 9 | ZS-6 | IC587370 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 10 | PTS-1 | IC587275 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 11 | PTS-5 | IC587279 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 12 | SHS-11 | IC587331 | Shopian | 33°9 | 74°99 | 5600ft |
| 13 | Cith W7 | IC587309 | Budgam | 33°54 | 75°07 | 5463 ft |
| 14 | BWS-19 | IC587311 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 15 | HS-4 | IC561062 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 16 | LG-4 | IC587077 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 17 | Cith W25 | IC587309 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 18 | Cith W11 | IC587310 | Anantnag | 33°73 | 75°16 | 5,253 ft |
| 19 | Cith W24 | IC587230 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 20 | BB-1 | IC587328 | Budgam | 33°54 | 75°07 | 5575ft |
| 21 | CWS-9 | IC587283 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 22 | CithW15 | IC587077 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 23 | PTS-10 | IC587286 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 24 | CWS-7 | IC587268 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 25 | BB-2 | IC587327 | Budgam | 33°54 | 75°07 | 5575ft |
| 26 | GWS-6 | IC587160 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 27 | BSS-4 | IC587298 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 28 | Cith W2 | IC561049 | Anantnag | 33°05 | 75°07 | 5414 ft |
| 29 | PBS-3 | IC587085 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 30 | SPS-1 | IC587172 | Shopian | 33°9 | 75°07 | 5414 |
| 31 | PTS-3 | IC587277 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 32 | PTS-2 | IC587276 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 33 | BB-3 | IC587326 | Budgam | 33°54 | 75°07 | 6991ft |
| 34 | PTS-21 | IC587334 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 35 | Cith W12 | IC587228 | Budgam | 34°63 | 76°04 | 10,479 ft |
| 36 | Cith W13 | IC587115 | Budgam | 34°63 | 76°04 | 10,479ft |
| 37 | Cith W23 | IC587091 | Anantnag | 33°73 | 75°15 | 5414ft |
| 38 | GB-2 | IC587072 | Ganderbal | 33°05 | 75°07 | 5414 ft |
| 39 | Cith W16 | IC587140 | Budgam | 34°63 | 76°04 | 10,479ft |
| 40 | BRTS-4 | IC587193 | Baramulla | 34°19 | 74°36 | 5,226 ft |
| 41 | Cith W5 | IC587086 | Pulwama | 33°88 | 74°92 | 5,350 ft |
| 42 | Cith W3 | IC587184 | Baramulla | 34°63 | 76°04 | 10,479ft |
| 43 | CithW14 | IC587091 | Anantnag | 33°73 | 75°15 | 5,253 ft |
| 44 | GLS-6 | IC587154 | Ganderbal | 34°23 | 74°78 | 5,312 ft |
| 45 | BRUS 10 | IC587218 | Baramulla | 34°19 | 74°36 | 5,226 ft |
| 46 | WGB-13 | Passport data send to NBPGR, New Delhi for IC number allotment Passport data send to NBPGR, New Delhi for IC number allotment | Budgam | 34°63 | 76°04 | 10,479 ft |
| 47 | CPB-4 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 48 | NB-2 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 49 | NDPB-1 | Baramulla | 34°19 | 74°36 | 5,226 ft | |
| 50 | PGB-1 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 51 | PB-3 | Pulwama | 33°88 | 74°92 | 5,350 ft | |
| 52 | PSB-2 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 53 | DKT-2 | Baramulla | 34°19 | 74°36 | 5,226 ft | |
| 54 | KD-2 | Baramulla | 34°19 | 74°36 | 5,226 ft | |
| 55 | NB-3 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 56 | AB-1 | Baramulla | 34°19 | 74°3 | 5226ft | |
| 57 | KD-1 | Baramulla | 34°19 | 74°36 | 5,226 ft | |
| 58 | UKB-3 | Baramulla | 34°19 | 74°36 | 5,226 ft | |
| 59 | JKB-5 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 60 | KB-1 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 61 | GGS-1 | Ganderbal | 34°23 | 74°78 | 5,312 ft | |
| 62 | ZP-3 | Pulwama | 33°88 | 74°92 | 5,350 ft | |
| 63 | BSS-5 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 64 | KB-2 | Baramulla | 34°19 | 74°36 | 5,226 ft | |
| 65 | BPM-2 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 66 | Shalimar-1 | Srinagar | 32°44 | 74°54 | 5,200ft | |
| 67 | GPB-2 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 68 | GKB-4 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 69 | CGB-1 | Budgam | 34°63 | 76°04 | 10,479ft | |
| 70 | ZGB-1 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 71 | GKB-2 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 72 | Shalimar-6 | Srinagar | 32°44 | 74°54 | 5,200ft | |
| 73 | NB-4 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 74 | DKB-1 | Budgam | 34°63 | 76°04 | 10,479ft | |
| 75 | S-289 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 76 | ZP-4 | Pulwama | 33°88 | 74°92 | 5,350 ft | |
| 77 | S-285 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 78 | Skua 0024 | Srinagar | 32°44 | 74°54 | 5,200ft | |
| 79 | Skua 0022 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 80 | Skua 003 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 81 | Skua 004 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 82 | DB-2 | Bandipora | 34°41 | 74°65 | 5541ft | |
| 83 | Skua 0023 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 84 | Skua 0020 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 85 | HCB-1 | Anantnag | 33°73 | 75°16 | 5,253 ft | |
| 86 | RKB-1 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 87 | GKB-1 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 88 | KBB-1 | Budgam | 34°63 | 76°04 | 10,479 ft | |
| 89 | Skua 002 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 90 | Sulaiman | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 91 | Saloora-1 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 92 | Wussan 8 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 93 | NBB-3 | Budgam | 34°63 | 76°04 | 10,479ft | |
| 94 | VL-2 | Ganderbal | 34°23 | 74°78 | 5,302 ft | |
| 95 | Chenovo | Exotic Collection | ||||
| 96 | Nugget | Exotic Collection |
DNA extraction
Total DNA was extracted from fresh, young leaf tissue (0.5 g), by a Cetyltrimethylammonium bromide (CTAB) method as described by Doyle [24]. The extract was further purified by standard method of Sambrook [25]. Concentration and purity of DNA was determined by electrophoresis in 0.8% agarose gels and spectrophotometrically on Nano Drop (Thermo). This extraction method yielded an average DNA content of 500 μg g-1 of leaf tissue, and purified DNA revealed A260/A280=1.80 ± 0.22, revealing the high purity of DNA samples.
RAPD and ISSR amplification
DNA from each walnut genotype was screened with 13 RAPD primers and 19 ISSR primers (Tables 2 and 3). For RAPD, the PCR reaction (50 μl) consisted of: 1X reaction buffer (20 mM Tris-HCl pH 8.4, 50 mM KCl), 0.2 mM dNTPs, 2 mM MgCl2, 10 pM primer, 1.0 unit of Taq DNA polymerase, and 25-50 ng genomic DNA. For ISSR the PCR reaction (25 μl) contained the following: 25 μL containing 40 ng genomic DNA, 1X PCR buffer (20 mM Tris-Cl pH 8.4, 50 mM KCl), 1.5 mM MgCl2, 0.2 mM dNTPs, 10 μM oligodeoxynucleotide primer and 1.0 unit of Taq DNA polymerase. PCR amplifications were performed in a gradient thermal cycler (Make TAKARA, Japan). The RAPD and ISSR amplified DNA was further analyzed by electrophoresis on 2% agarose gel in a 0.5x TBE buffer. The gels were stained with ethidium bromide (0.5 μg/ml) and visualized under UV light. A 100 bp DNA ladder (Invitrogen, Madison, USA) was used for molecular weight estimation of PCR products. Only the bands between 100 and 1500 bp which were clear and unambiguous were recorded.
Table 2: Polymorphic profile and characteristics of 13 RAPD primers used across 96 genotypes of walnut (Juglans regia L.).
| S. No. | ISSR | TB | PB | PIC | IB | MI | Primer Sequence |
|---|---|---|---|---|---|---|---|
| 1 | 834 | 4 | 4 | 0.25 | 6.06 | 1.00 | (AG)8GC |
| 2 | 849 | 3 | 2 | 0.26 | 3.56 | 0.77 | (GT)7GAA |
| 3 | 811 | 9 | 9 | 0.42 | 9.65 | 3.77 | CAC(CA)6AT |
| 4 | 818 | 3 | 3 | 0.28 | 4.33 | 0.84 | (CA)8G |
| 5 | 821 | 2 | 2 | 0.44 | 2.65 | 0.87 | (GT)8T |
| 6 | 822 | 3 | 3 | 0.37 | 2.58 | 1.11 | (TC)7TAC |
| 7 | 826 | 4 | 4 | 0.22 | 4.79 | 0.88 | (AC)8C |
| 8 | 827 | 2 | 2 | 0.16 | 3.60 | 0.33 | (AC)8GG |
| 9 | 828 | 3 | 3 | 0.24 | 4.31 | 0.73 | (TG)8A |
| 10 | 845 | 2 | 2 | 0.28 | 2.98 | 0.56 | (CT)8GT |
| 11 | 864 | 2 | 2 | 0.20 | 3.52 | 0.39 | (ATG)5TG |
| 12 | 865 | 6 | 6 | 0.37 | 7.67 | 2.24 | (ATG)5TCC |
| 13 | 819 | 1 | 1 | 0.12 | 1.88 | 0.12 | (GT)8A |
| 14 | 857 | 4 | 4 | 0.35 | 2.90 | 1.40 | (AC)8GGTC |
| 15 | 859 | 6 | 6 | 0.31 | 3.75 | 1.83 | (TG)8RC |
| 16 | 848 | 6 | 5 | 0.26 | 6.33 | 1.58 | (CA)8GC |
| 17 | 830 | 4 | 1 | 0.04 | 6.15 | 0.17 | (TG)8G |
| 18 | 851 | 7 | 7 | 0.22 | 2.06 | 1.54 | (GT)8CG |
| 19 | 847 | 1 | 1 | 0.49 | 1.10 | 0.49 | (CA)8GC |
Table 3: Polymorphic profile and characteristics of 19 ISSR primers used across 96 genotypes of walnut (Juglans regia L.)
| S. No. | ISSR | TB | PB | PIC | IB | MI | Primer Sequence |
|---|---|---|---|---|---|---|---|
| 1 | 834 | 4 | 4 | 0.25 | 6.06 | 1.00 | (AG)8GC |
| 2 | 849 | 3 | 2 | 0.26 | 3.56 | 0.77 | (GT)7GAA |
| 3 | 811 | 9 | 9 | 0.42 | 9.65 | 3.77 | CAC(CA)6AT |
| 4 | 818 | 3 | 3 | 0.28 | 4.33 | 0.84 | (CA)8G |
| 5 | 821 | 2 | 2 | 0.44 | 2.65 | 0.87 | (GT)8T |
| 6 | 822 | 3 | 3 | 0.37 | 2.58 | 1.11 | (TC)7TAC |
| 7 | 826 | 4 | 4 | 0.22 | 4.79 | 0.88 | (AC)8C |
| 8 | 827 | 2 | 2 | 0.16 | 3.60 | 0.33 | (AC)8GG |
| 9 | 828 | 3 | 3 | 0.24 | 4.31 | 0.73 | (TG)8A |
| 10 | 845 | 2 | 2 | 0.28 | 2.98 | 0.56 | (CT)8GT |
| 11 | 864 | 2 | 2 | 0.20 | 3.52 | 0.39 | (ATG)5TG |
| 12 | 865 | 6 | 6 | 0.37 | 7.67 | 2.24 | (ATG)5TCC |
| 13 | 819 | 1 | 1 | 0.12 | 1.88 | 0.12 | (GT)8A |
| 14 | 857 | 4 | 4 | 0.35 | 2.90 | 1.40 | (AC)8GGTC |
| 15 | 859 | 6 | 6 | 0.31 | 3.75 | 1.83 | (TG)8RC |
| 16 | 848 | 6 | 5 | 0.26 | 6.33 | 1.58 | (CA)8GC |
| 17 | 830 | 4 | 1 | 0.04 | 6.15 | 0.17 | (TG)8G |
| 18 | 851 | 7 | 7 | 0.22 | 2.06 | 1.54 | (GT)8CG |
| 19 | 847 | 1 | 1 | 0.49 | 1.10 | 0.49 | (CA)8GC |
Molecular Data Analysis
For qualitative analysis, the RAPD and ISSR data were analyzed as dominant markers. Presence or absence of the band was scored as 1 or 0 respectively, thus obtaining the molecular identification profile for each genotype. The capacity of each primer to distinguish among the genotypes was evaluated by the number of amplicons per primers, Percent Polymorphism, Polymorphic Information Content (PIC) [26], Effective Multiplex Ratio (EMR), Resolving Power (Rp) [27] and Marker Index (MI) [28].
The dendrogram was constructed using NTSYS-pc version 2.02e software [29] (Applied BioStatistics, Inc., Setauket, NY, USA) package to compute pairwise Jaccard’s similarity coefficients [30]and the similarity matrix thus obtained was used in cluster analysis using an UPGMA (unweighted pair-group method with arithmetic averages) and sequential, agglomerative, hierarchical and nested (SAHN) clustering algorithm. In order to estimate the goodness of fit of the UPGMA cluster analysis between similarity matrix and the dendrogram of RAPD and ISSR, the coefficient of correlation was calculated using the Mantel test [31] using the MxComp Module of NTSYS PC software (version 2.02e) (Applied BioStatistics, Inc., Setauket, NY, USA) package.
Genic Variation and Heterozygosity Statistics of 13 RAPD and 19 ISSR primers were calculated using software-PopGene-32 version 1.31 [32].
Results and Discussion
RAPD analysis
Thirteen RAPD primers used in present study produced 54 scorable fragments out of which 15 (27.78%) were polymorphic across 96 walnut genotypes. The number of polymorphic alleles found at each RAPD locus ranged from 0 to 7 with an average of 1.15 loci per primer. The most prolific RAPD primer was OPC-04 that amplified 7 polymorphic loci. Significantly higher level of polymorphism (43%) was observed in walnut through RAPD markers by Fakhraddin [33]. Our study revealed higher number of average alleles per loci, on an average 4.15 alleles was observed per loci which are higher than previous reports of Nicese [11] and Potter [34]. Resolving power (Rp) values for RAPD primers ranged from 0 to 12 with an average of 6.99. MI value for RAPD ranged from 0 to 1.36 with an average of 0.32. The PIC values of the primer combinations in RAPD varied from 0 to 0.29 with an average of 0.07. Our average values of MI and RP are higher than those which were observed by Fatahi [35] and Ahmed [17]. The respective values for overall genetic variability for PIC, Rp and MI in all the 96 genotypes are summarized in Table 2. The RAPD derived data was subjected to calculate the genetic similarity which revealed a varying degree of genetic relationship for genotypes belonging to different geographically isolated districts. The Jaccard’s similarity coefficient ranged from 0.78 to 1.00 with an average of 0.91 amongst all the 96 walnut genotypes. The lowest pair-wise similarity coefficient value was between CITH-W3 and PTS-10 (0.78). This indicates a quite high degree of genetic variability within the species which may be because of different original parents. Cluster analysis defined four main groups (Figure 1). First Cluster consisted of 26 genotypes with an average similarity of 94%. Second cluster, the major cluster comprised of 45 genotypes with an average similarity of 95%. Third and Fourth Cluster consisted of 4 and 21 genotypes respectively with an average similarity of 89% and 93% respectively. The principal coordinate analysis (PCoA) showed that the first three axis accounted for 94.59% (91.36%, 2.12% and 1.08% by 1st, 2nd and 3rd co-ordinate respectively) of total variation. The grouping shown in dendrogram was at par with that shown in 3D scatter of PCoA (Figure 2).
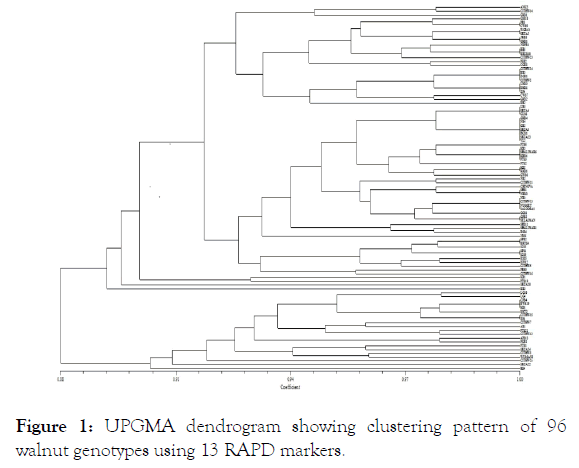
Figure 1: UPGMA dendrogram showing clustering pattern of 96 walnut genotypes using 13 RAPD markers.
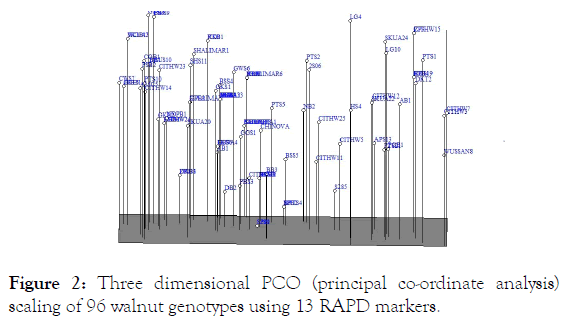
Figure 2: Three dimensional PCO (principal co-ordinate analysis) scaling of 96 walnut genotypes using 13 RAPD markers.
The number of alleles (na), the effective number of alleles (ne) [36], Nei’s gene diversity (h) and Shannon's Information index (I) [37] are given in Table 4. The average effective number of alleles for all the RAPD primers was 0.76 with locus OPU-03 possessing highest number of effective alleles (1.48) followed by OPJ-04 (0.45) and OPC-04 (0.27). Similarly, Shannon's Information index ranged from 0 to 0.43 (OPU-03) with an average of 0.12 and Nei’s gene diversity ranged from 0 to 0.29 (OPU-03) with an average of 0.08.
Table 4: Genetic Variation Statistics of 13 RAPD Primers in 96 Walnut genotypes.
| S. No. | Primer Name | na* | ne* | h* | I* |
|---|---|---|---|---|---|
| 1 | OPC-04 | 2.00 | 1.27 | 0.19 | 0.32 |
| 2 | OPB-01 | 1.00 | 1.00 | 0.00 | 0.00 |
| 3 | OPJ-04 | 1.50 | 1.45 | 0.24 | 0.33 |
| 4 | OPM-5 | 1.20 | 1.14 | 0.08 | 0.12 |
| 5 | OPM-09 | 1.00 | 1.00 | 0.00 | 0.00 |
| 6 | OPO-02 | 1.33 | 1.32 | 0.16 | 0.23 |
| 7 | OPP-03 | 1.00 | 1.00 | 0.00 | 0.00 |
| 8 | OPR-03 | 1.00 | 1.00 | 0.00 | 0.00 |
| 9 | OPU-03 | 1.75 | 1.48 | 0.29 | 0.43 |
| 10 | OPB-08 | 1.17 | 1.04 | 0.03 | 0.06 |
| 11 | OPC-06 | 1.17 | 1.02 | 0.02 | 0.04 |
| 12 | OPM-02 | 1.00 | 1.00 | 0.00 | 0.00 |
| 13 | OPP-07 | 1.00 | 1.00 | 0.00 | 0.00 |
| Mean | 1.28 | 1.12 | 0.08 | 0.12 | |
| St. Dev | 0.45 | 0.26 | 0.15 | 0.22 |
*na=Observed number of alleles; *ne=Effective number of alleles [Kimura and Crow (1964)]; *h=Nei's (1973) gene diversity; *I=Shannon's Information index [Lewontin (1972)].
ISSR analysis
ISSR markers are helpful in fields of genetic diversity, phylogenetic studies, gene tagging, genome mapping and evolutionary biology in a wide range of plant species [6]. They have been effectively used in many tree species, including walnut (J. regia L.) [34,38], olive (Olea europaea L.) [39], mulberry (Morus L.) [40] and plum (Prunus L. spp.) [41]. The genetic variation observed through RAPD was further analyzed by ISSR to obtain a more accurate molecular characterization of the studied walnut genotypes. ISSR analysis of 19 markers produced 72 scorable fragments out of which 67 (93.05%) were polymorphic. The number of alleles observed at each ISSR locus varied from 1(p819; p847) to 9 (p811) with an average of 3.78 alleles per locus and polymorphic loci also range from 1 (p 847; p819; p830) to 9 (p811) with an average of 3.52 polymorphic loci per primer. The most prolific ISSR primer was p811 amplifying 9 polymorphic bands, while the least productive markers amplifying single monomorphic loci were p819 and p847. Hence we observe that ISSR markers have higher potential to discriminate walnut genotypes from each other due to capability of generating higher number of polymorphic loci per primer. PIC values range from 0.04 (p830) to 0.49 (p847) with an average of 0.27. MI values ranged from 0.12 (p830) to 3.77 (p847) with an average of 1.08. Primer p811 exhibited the highest Rp value (9.64) and the lowest value (1.10) was recorded for the primer p847 with an average of 4.20. Prevost and Wilkinson (1999) reported a linear relationship between the ability of a primer to distinguish genotypes and Resolving power. The percentage of polymorphic bands (PPB), the average number of polymorphic bands per primer, PIC, MI and Rp obtained in our present results were higher than those previously found in ISSR studies in walnut by Potter [34] Christopoulus [12] Mahmoodi [13] and Ji [42].
The Jaccard’s similarity coefficient ranged from 0.22 to 0.85 with an average of 0.58 among all the 96 walnut genotypes used which is lower than the findings of Ji [43]. The lowest pair-wise similarity (22%) was observed between genotype HCB1 and Chinovo while as highest pair-wise similarity (85%) was observed between genotype DKT2 and BWS19. The reason for this higher similarity is that the two genotypes may have the same parents. In the present study, 94 genotypes were separated into ten distinct clusters and two genotypes (HCB1 and PSB2) do not form part of any cluster (Figure 3). First cluster consisted of 2 genotypes with an average similarity of 58.1%. Second cluster, the largest one, consisted of 25 genotypes with an average similarity of 68.1%. Third, fourth and fifth cluster consisted of 21, 13 and 2 genotypes with an average similarity of 68.6%, 62.4% and 63.5% respectively. Similarly sixth, seventh and eighth cluster was represented by 11, 6 and 2 genotypes with an average similarity of 66.4%, 66% and 62.7% respectively. Ninth and tenth cluster comprised of 10 and 2 genotypes with an average similarity of 63% and 60.5% respectively.
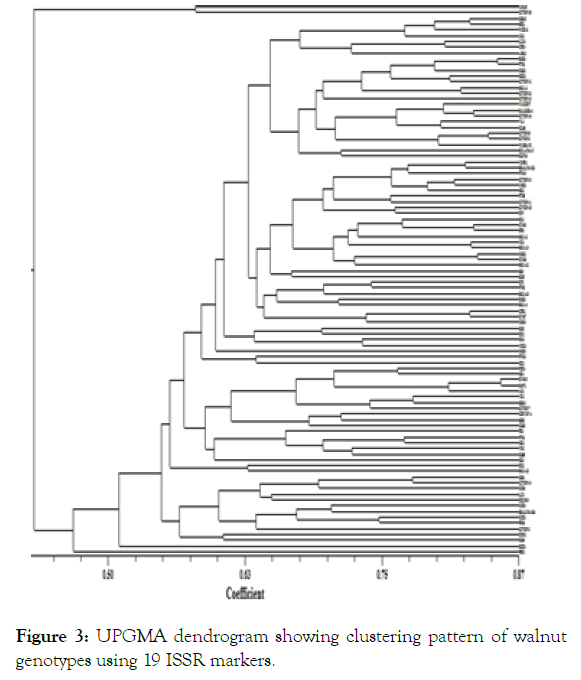
Figure 3: UPGMA dendrogram showing clustering pattern of walnut genotypes using 19 ISSR markers.
Furthermore individuals of two main clusters (Cluster 2 and Cluster 3) have an average similarity of 65.9%. The principal coordinate analysis (PCoA) showed that the first three axis accounted for 65.65% (59.39%, 3.34% and 2.90% by 1st, 2nd and 3rd co-ordinate respectively) of total variation. The grouping shown in dendrogram was at par with that shown in 3D scatter (Figure 4). The na, ne, h and I are summarized in Table 5. The locus p847 possessed highest number (1.98) of effective alleles while locus p830 possessed lowest number (1.05) of effective alleles with an average of 1.48 for all the studied loci. Similarly, Shannon's Information index ranged from 0.07 (p830) to 0.69 (p847) with an average of 0.44 and Nei’s gene diversity ranged from 0.04 (p830) to 0.49 (p847) with an average of 0.29. Ji et al. recorded a high genetic diversity as detected with Nei’s (H=0.25) and Shannon’s diversity (I=0.40) which is at par with our findings.
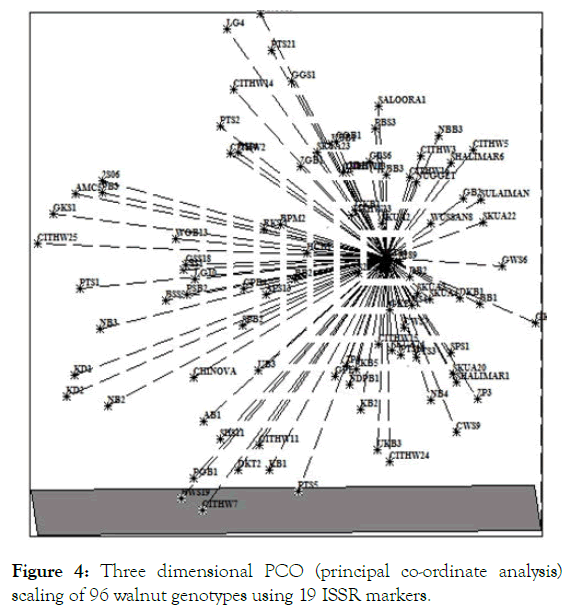
Figure 4: Three dimensional PCO (principal co-ordinate analysis) scaling of 96 walnut genotypes using 19 ISSR markers.
Table 5: Genetic variation statistics of 19 ISSR Primers in 96 Walnut genotypes.
| S. No. | Primer name | na* | ne* | h* | I* |
|---|---|---|---|---|---|
| 1 | p834 | 2.00 | 1.38 | 0.25 | 0.40 |
| 2 | p849 | 1.67 | 1.44 | 0.26 | 0.38 |
| 3 | p811 | 2.00 | 1.74 | 0.42 | 0.61 |
| 4 | p818 | 2.00 | 1.45 | 0.28 | 0.44 |
| 5 | p821 | 2.00 | 1.79 | 0.44 | 0.63 |
| 6 | p822 | 2.00 | 1.65 | 0.37 | 0.55 |
| 7 | p826 | 2.00 | 1.36 | 0.22 | 0.36 |
| 8 | p827 | 2.00 | 1.23 | 0.16 | 0.27 |
| 9 | p828 | 2.00 | 1.38 | 0.24 | 0.38 |
| 10 | p845 | 2.00 | 1.53 | 0.28 | 0.42 |
| 11 | p864 | 2.00 | 1.28 | 0.20 | 0.33 |
| 12 | p865 | 2.00 | 1.64 | 0.37 | 0.56 |
| 13 | p819 | 2.00 | 1.13 | 0.12 | 0.23 |
| 14 | p857 | 2.00 | 1.61 | 0.35 | 0.52 |
| 15 | p859 | 2.00 | 1.50 | 0.31 | 0.47 |
| 16 | p848 | 1.83 | 1.44 | 0.26 | 0.40 |
| 17 | p830 | 1.25 | 1.05 | 0.04 | 0.07 |
| 18 | p851 | 2.00 | 1.34 | 0.22 | 0.36 |
| 19 | p847 | 2.00 | 1.98 | 0.49 | 0.69 |
| Mean | 1.93 | 1.48 | 0.29 | 0.44 | |
| St. Dev | 0.26 | 0.35 | 0.17 | 0.22 |
*na=Observed number of alleles; *ne=Effective number of alleles [Kimura and Crow (1964)]; *h=Nei's (1973) gene diversity; *I=Shannon's Information index [Lewontin (1972).
RAPD and ISSR combination
For obtaining more desirable results, the data of both the primer types (RAPD & ISSR) was pooled and a dendrogram was developed from this data. The Jaccard’s similarity coefficient based on all possible pairs of genotypes varied from 0.59 (SKUA-22 & CITH-W-25) to 0.92 (DKT-2 & BWS-19) with an average of 0.74. The UPGMA dendrogram clearly depicted the genetic relationship between the genotypes and showed that all the genotypes formed one major cluster and five minor clusters and one genotype PTS21 does not form the part of any cluster. Cluster I, the major cluster, comprising of 63 genotypes with 77% average similarity represented the selections from Budgam (31.7%), Ganderbal (30.15%), Pulwama (11.11%), Anantnag (6.34%), Srinagar (4.76) and Shopian (3.17%). Thus the majority of genotypes in Cluster I are from district Budgam followed by Ganderbal. Within the first cluster, two subgroups consisting of 52 and 11 genotypes were found. The second cluster comprised of only two genotypes each from Budgam and Ganderbal district with an average similarity of 78%. The third cluster comprising of eleven genotypes with an average similarity of 77% represented the genotypes from district Budgam (45.45%), Pulwama (18.18%) and 9.0% each from district Ganderbal, Baramulla, Shopian and exotic collections. Therefore the majority of genotypes in this cluster are from district Budgam. Fourth cluster consisted of 7 genotypes with an average similarity of 81%. This cluster was represented by genotypes from Budgam (42.85%) and Baramulla (42.85%). Fifth cluster consisted of 9 genotypes with an average similarity of 76%. This cluster was represented by the genotypes from district Ganderbal (33.35%), Budgam (22.2%), Anantnag (22.2%), Pulwama (11.1%) and Baramulla (11.1%). Sixth cluster consisted of 3 genotypes each from district Ganderbal, Budgam and Anantnag with an average similarity of 76%. The outcrossing nature and seedling origin of walnut may be responsible for such diverse clustering. No relationship was found between the spatial and genetic proximity of the germplasm apart from few genotypes which may be originated from similar regions. Since the genotypes have seedling origin and were grown as nuts which shows high degree of segregation and thus leading to development of entirely new genotypes. Thus the genotypes showing similarity may be derived from the genotypes from the same cluster. The common practice in the region was to raise the walnut trees from nuts of genotypes having superior quality and thus leading to development of new walnut genotypes with some degree of superior nut qualities. The principal coordinate analysis (PCoA) showed that first three axis accounted for 78.53% (74.82, 2.01 and 1.69 by Ist, 2nd and 3rd co-ordinate respectively) of total genetic variance. The grouping shown in dendrogram was at par with that shown in 3D scatter.
Two way Mantel test [31] was done between the RAPD and ISSR data matrices to compare the goodness of the fit of each similarity matrix. The correlation coefficient (r) was found to be 0.11 between matrices generated by RAPD and ISSR markers using the Mantel Test (t=2.39, p=0.99) supporting the validity of the dendrogram.
Both the marker techniques of RAPD and ISSR proved to be extremely effective in discriminating the 96 walnut genotypes analyzed, since the majority of bands were polymorphic amongst genotypes. The number and percent of polymorphic loci, polymorphic information content (PIC), and marker index (MI) of primers were higher for ISSR markers. In this study, we found a high degree of polymorphism (93.05%) in the 96 genotypes studied. This validates that ISSR markers are useful markers in genetic divergence studies allowing an unequivocal identification for genotypes. The high level of polymorphism possibly reflects the outcrossing character of walnut because almost similar results have been obtained in fruit and nut tree species like pistachio [42] or olive [44].
Conclusion
Present study provides the base for association mapping studies and linkage map development in walnut. Since the walnut population we studied is heterogeneous with respect to number of traits and those traits can be linked with markers through association mapping studies. Present study clearly identified the markers with potential for utilization in walnut breeding programs.
Acknowledgements
The authors express their gratitude to Central Institute of Temperate Horticulture (Indian Council of Agricultural Research, ICAR) and University Grants Commission (UGC) for financial support of this work.
REFERENCES
- McGranahan GH, Charles A, Leslie CA, Philips HA, Dandaker A. Walnut propagation, in: Walnut production manual (Romos D, ed). University of California, Davis. 1998;pp.71-83.
- INC. Global Statistical Review. 2014-2015. 2015;pp. 1-76.
- Arranz S, Cert R, Pérez-Jiménez J, Cert A, Saura-Calixto F. Comparison between free radical scavenging capacity and oxidative stability of nut oils. Food Chem. 2008;110:985-990.
- Rahimipanah M, Hamedi M, Mirzapour M. Antioxidant activity phenolic contents of Persian walnut (Juglans regia L.) green husk extract. AJFST. 2010;1:105-111.
- Ros E, Mataix J. Fatty acid composition of nuts–implications for cardiovascular health. Br J Nutr. 2006;96:29-35.
- Jabbarzadeh Z, Khosh-khui M, Salehi H, Saberivand A. Inter simple sequence repeat (ISSR) markers as reproducible and specific tools for genetic diversity analysis of rose species. Afr J Biotechnol. 2010;9:6091-6095.
- Francesca PI, Pamfil D, Raica P, Petricele. Assessment of the genetic variability among some Juglans cultivars from the Romanian National Collection at SCDP vâlcea using RAPD markers. Rom Biotech Lett. 2010;15:41-49.
- Cubero J. Introduction to plant genetic improvement. Mundi-Press editions, Madrid. 2003.
- Ebrahimi A, Fatahi R, Zamani Z. Analysis of genetic diversity among some Persian walnut genotypes (Juglans regia L.) using morphological traits and SSRs markers. Sci Hortic. 2011;130:146-151.
- Fjellstrom RG, Parfitt DE, Mvgranahan GH. Genetic relationship and characterization of Persian walnut (Juglans regia L.) cultivars using restriction fragment length polymorphism (RFLPs). J Amer Soc Hort Sci. 1994;119:833-839.
- Nicese FP, Hormaza JI, McGranahan GH. Molecular characterization genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers. Euphytica. 1998;101:199-206.
- Christopoulos MV, Rouskas D, Tsantili E, Bebeli PJ. Germplasm diversity and genetic relationships among walnut (Juglans regia L.) cultivars and Greek local selections revealed by inter-simple sequence repeat (ISSR) markers. Sci Hortic. 2010;125:584-592.
- Mahmoodi R, Rahmani F, Paktarmani R. Genetic diversity of Persian walnut from Iran as revealed by intersimple sequence repeat (ISSR) markers. J Am Pomol Soc. 2012;66:101-106.
- Foroni I, Woeste K, Monti ML, Rao R. Identification of ‘Sorrento’ walnut using simple sequence repeats (SSRs). Genet Resour Crop Ev. 2007;54:1081-1094.
- Bayazit S, Kazan K, Gülbitti S, Çevik V, Ayanoğlu H. AFLP analysis of genetic diversity in low chill requiring walnut (Juglans regia L.) genotypes from Hatay, Turkey. Sci Hortic. 2007;111:394-398.
- Solar A, Podjavorsek A, Stamar F. Phenotypic and genotypic diversity of European chestnut (Castanea sativa Mill.) in Slovenia-opportunity for genetic improvement. Genet Resour Crop Ev. 2005;52:381-394.
- Ahmed N, Mir JI, Mir RR, Rather NA, Rashid R. SSR and RAPD analysis of genetic diversity in walnut (Juglans regia L.) genotypes from Jammu and Kashmir, India. Physiol Mol Biol Plants. 2012;18:149-160.
- Wani N, Ahmad MF, Bhat MA, Wani SA, Iqbal J. Diversity analysis in walnut (L.) using morphological and RAPD markers. Green Farming. 2014;4:533-537.
- Pérez de la Torre MC, Escandón A. Construction of a molecular identification profile of new varieties of Nierembergia linariaefolia by anchored microsatellites. Electron J Biotechn. 2006;9:248-252.
- Escandón A, Zelener N, Pérez de la Torre M, Soto S. Molecular identification of new varieties of Nierembergia linariaefolia (Graham), a native Argentinean ornamental plant. J Appl Genet. 2007;48:115-123.
- Martins-Lopes P, Lima-Brito J, Gomes SN, Meirinhos J, Santos L. RAPD and ISSR molecular markers in Olea europaea L.: Genetic variability and molecular cultivar identification. Genet Resour Crop Ev. 2007;54:117-128.
- Li S, Li J, Yang X, Cheng Z, Zhang W. Genetic diversity and differentiation of cultivated ginseng (Panax ginseng C.A. Meyer) populations in North-east China revealed by inter-simple sequence repeat (ISSR) markers. Genet Resour Crop Ev. 2010;58:815-824.
- Kamalesh SM, Tikam SR, Shirish AR. Molecular analyses of genetic variability in soap nut (Sapindus mukorossi Gaertn). Ind Crop Prod. 2011;34:1111-1118.
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13-15.
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Habor Lab Press, Cold Spring Harbor, New York. 1989;1-34.
- Weising K, Nybon H, Wolff K, Kahl G. Applications of DNA fingerprinting in plant sciences. in: Taylor & Francis eds. DNA Fingerprinting in Plants. Principles, Methods and Applications (2nd edition). 2005;pp.235-276.
- Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 1999;98:107-112.
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed. 1996;2:225-238.
- Rohlf FJ. NTSYS-pc numerical taxonomy and multivariate analysis system, vers. 2.00. Exeter Software, Setauket, New York. 1998;1-43.
- Jaccard P. New research on floral distribution. Bull Soc Vaud Sci Nat. 1908;44:223-270.
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209-220.
- Yeh F, Yang RC, Boyle T. Genic variation and heterozygosity statistics software-popgene-32 version 1.31. University of Alberta, Edmonton, Canada. 1999.
- Fakhraddin MHS, Nawroz ART, Jamal MF. Assessment of genetic relationship among some iraqi walnut genotypes (Juglans regia L.) in Sulaimani Region Using RAPD and SSR Molecular Markers. JJAS. 2013;9:351-362.
- Potter D, Gao F, Aiello G, Leslie C, McGranahan G. Intersimple sequence repeat markers for fingerprinting and determining genetic relationships of walnut (Juglans regia) Cultivars. J Am Soc Hortic Sci. 2002;127:75-81.
- Fatahi R, Ebrahimi A, Zamani Z. Characterization of some iranians and foreign walnut genotypes using morphological traits and RAPD Markers. Hort Environ Biotechnol. 2010;51:51-60.
- Kimura M, Crow J. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725-738.
- Lewontin RC. The apportionment of human diversity. Evol Biol. 1972;6:381-398.
- Pollegioni P, Bartoli S, Cannata F, Malvolti ME. Genetic differentiation of four Italian walnut (Juglans regia L.) varieties by inter simple sequence repeat (ISSR). J Genet Breed. 2003;57:231-240.
- Terzopoulos PJ, Kolano B, Bebeli PJ, Kaltsikes PJ, Metzidakis I. Identification of Olea europaea L. cultivars using inter-simple sequence repeat markers. Sci Hort. 2005;105:45-51.
- Vijayan K, Srivastava PP, Nair CV, Tikader A, Awasthi AK. Molecular characterization and identification of markers associated with leaf yield traits in mulberry using ISSR markers. Plant Breed. 2006;125:298-301.
- Liu W, Li S, Zhang A, Liu D. Genetic diversity revealed by RAPD markers in plum collection of China. Acta Hortic. 2007;734:287-294.
- Ji A, Wang Y, Wu G, Wu W, Yang H. Genetic diversity and population structure of North China mountain walnut revealed by ISSR. AJPS. 2014;5:3194-3202.
- Hormaza JI, Dollo L, Polito VS. Determination of relatedness and geographical movement of Pistacia vera (pistachio; Anacardiaceae) germplasm by RAPD analysis. Econ Bot. 1994;48:349-358.
- Fabbri A, Hormaza JI, Polito VS. Random amplified polymorphic DNA analysis of olive (Olea europaea L.) cultivars. J Amer Soc Hort Sci. 1995;120:538-542.
Citation: Fazili KM, Mir JI, Ahmed N, Fazili KM (2019) Genetic Diversity Analysis of Walnut (Juglans regia L.) from Kashmir Valley Using RAPD and ISSR Markers. Agrotechnology 8:185. doi: 10.35248/2168-9881.19.8.185
Copyright: © 2019 Fazili KM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


