Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- RefSeek
- Hamdard University
- EBSCO A-Z
- SWB online catalog
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 13, Issue 4
Four Chromosomally Clustered Amph Class-C β-Lactamases Control the Cephalosporin Hydrolysis in Mycobacterium tuberculosis and Similar Genetic Loci Appeared as Pseudogenes in Mycobacterium leprae
Asit Kumar Chakraborty*Received: 13-Sep-2023, Manuscript No. PDS-23-22979; Editor assigned: 15-Sep-2023, Pre QC No. PDS-23-22979 (PQ); Reviewed: 29-Sep-2023, QC No. PDS-23-22979; Revised: 13-Dec-2023, Manuscript No. PDS-23-22979 (R); Published: 20-Dec-2023, DOI: 10.35250/2167-1052.24.13.374
Abstract
Mycobacterium tuberculosis (Mt) chromosomal ten PBPs and one class-A β-lactamase (blaC) were implicated in multidrug resistance against penicillin, cephalosporin and carbapenem drugs. The Mt PonA1 and PonA2 had 34% homologies to E. coli class-A (PBP1A and PBP1B) proteins whereas E. coli class-B PBP2 referred as PbpA and PbpB sub-class proteins with 27% homologies in Mt. The class-C PBPs were, PBP4, MecA_N, DacB1, DacB2 and AmpH1- AmpH4. The PBP3 and PBP5 of E. coli were missing in Mt although PBPB called as PBP3 of Mt whereas AmpH designated sometime as DacA or PBP5. Similarly, AmpH protein wrongly called PBP4 where we could not find any similarity to E. coli PBP4. We investigated the Mt whole genomes (accession nos. AL123456, CP001642, CP054013, CP001641, CP025597) to get authentic PBP4 in Mt strain FDAARGOS_757 genome (protein id. AUP69687, 34% homology to E. coli PBP4) which was designated as conserved protein in chromosomes of Mt strains H37Rv, H37Rv-1, GG-36-11 and CCDC5180 making confusion in data analysis. Similarly, we BLASTP homology searched with plasmid-mediated 24 β-lactamases to confirm four blaAmpH genes, (AmpH1, AmpH2, AmpH3 and AmpH4) that predominantly controlled the degradation of cephalosporins in M. tuberculosis but such genes were found as pseudogenes in M. leprae. The Mt AmpH1/2/3/4 enzymes had better homologies with V. parahaemolyticus and Yersinia pekkanenii AmpH enzyme as well as with E. coli AmpH enzyme. The DacB1 (PBP6) and DacB2 (PBP7) enzymes had blaTEM similarity but no AmpH similarity suggesting such PBPs and BlaC β-lactamase controlled the penicillin hydrolysis. The blaC enzyme had 30% homology to S. aureus blaZ β-lactamase but such enzyme was also missing in M. leprae. Homology search suggested carbapenem hydrolysis by blaOXA-23/51-like PBPA/B enzymes and blaOXA-58 related MecA_N domain β-lactamase which has 21% similarity to S. aureus mecA enzyme. The PonA1/A2 had no homology to 24 classes of β-lactamases but still were popular enzymes implicated for better penicillin hydrolysis. We designed primers for Mt PBPs to check transcription of individual genes by RT-PCR as well as to check chromosomal locus by BLASTN search after WGS. The E. coli genome had no similarities to blaAmpH1/2/3/4 genes but a 17nt (5’-CCTTGGTGCCGTCGACC-3’) sequence found in pKEC-a3c plasmid of C. fruendii with homology to blaAmpH4 gene (nt. 1236-1252) and shared a homology with IS1182-like ISKpn6 transposase of plasmids. The oligonucleotides selected many Mt chromosomes and located conserved blaAmpH4 protein in all cases. However, D435E and F495V mutations of blaAmpH4 in the Mt strain 5521 were evident and frameshift deletions located in Mt stain FDAARGOS_756 blaAmpH4 gene with no protein was made. Thus, mutations, deletions and rearrangements mediated by IS-elements were the driving force to make new AmpH genes during long time TB treatment with 4-6 drugs.
Keywords
PBP4; blaAmpH; blaC; TB; Cephalosporin; Leprosy; ISKpn transposase; BLAST-2; AmpH primers
Abbreviations
AN: Accession Number; Mt: Mycobacterium tuberculosis; BLAST: Basic Local Alignment Search Tool; RTPCR: Reverse Transcription Polymerase Chain Reaction; WGS: Whole Genome Sequencing; PBP: Penicillin-Binding-Protein
Introduction
The estimated 1.9 million deaths from tuberculosis in 2020 [1]. The COVID-19 pandemic between 2020-2022 highly impacted TB treatment similar to HIV pandemic earlier [2]. In 2023, 4.5 million TB infections is a concern for India. The proper diagnosis is important for any disease to know its progress in the population. TB bacilli from patients hardly grow in medium-agar plate quickly and thus drug sensitivity test is not possible. Eventually, all TB patients have to take 4-6 different drugs every day for at least 2 months and then 2-3 drugs for more 4 months [3-5]. Whatever the case, PENEM drugs were not popular for the treatment of TB although some popular high growing Mycobacterium strains were very sensitive to imipenem and meropenem [6-10]. The early TB cases (1940-1960) were easily cured by rifampicin and streptomycin drugs but rpoB, rrs, rpsL genes mutation caused drug resistance. The rifampicin inhibits RNA polymerase enzyme by binding to it and RNA synthesis is blocked while β-subunit (rpoB gene) mutation gives resistance [11,12]. Some acetyl-and phospho-transferases also caused streptomycin and aminoglycoside resistant due to acetylation and phosphorylation of the antibiotics which were protein synthesizing inhibitors [13,14]. The fluroquinolone drugs like ciprofloxacin, norfloxacin and moxifloxacin were also helpful to clear TB infection but DNA gyrase gene (gyrA/gyrB subunits) mutations caused drug resistance in many cases [15]. Macrolide antibiotics erythromycin, kanamycin and higher derivative amikacin were still used for the TB treatment but MmpL5, MmpL7, Stp, JefA, macrolide drug efflux enzymes and erythromycin esterase were implicated as their resistance [16-20]. Thus, all conventional drugs may not be helpful today and TB specific drugs like isoniazid, dapsone, pyrazinamide, ethambutol and linezolid were prescribed although some resistance to those very important drugs were already noticed through WGS sequencing [21-27]. In truth, no plasmid has been discovered in M. tuberculosis (Mt). Since 2015 few plasmids have been sequenced from other high replicating Mycobacterium strains but hardly found any MDR gene [28,29]. Careful plasmid isolation from 6 ml overnight culture may be good to see any plasmid in Mt [30-32]. In an elegant experiment with Indian TB patients and their family, it was concluded that nutritional food supplement caused rapid cure of TB patients as well as reduced the infections to other family members. Thus, we undertake to find the cause of penicillin drug resistance in TB and also want to find the cause of dissimilarity between Mt PBP4 verses E. coli PBP4 as we have seen in our study. We collected the all PBPs or β-lactamases from Mt genome by naked eye (5 hrs/genome; 4.4 million bases) and then compared with E. coli PBP1A, PBP1B, PBP2, PBP3, PBP4, PBP5, PBP6, PBP7, AmpH and class-A, class-C and class-D β-lactamases like blaTEM, blaSHV, blaCTXM, blaOXA-23, blaOXA-58, blaAmpC, blaKPC, blaDHA and blaNDM-1 to classify Mt PBPs and β-lactamases [33-36].
First antibiotic discovered in 1926 is penicillin from fungus Penicillium notatam through the process of fermentation [37]. Sadly, a penicillinase reported as early as 1940 and the said gene (AMP) was detected in plasmid pBR322 during 1960's [38]. Further appearance of blaTEM, blaSHV, blaOXA penicillinases, scientist developed cephalosporin antibiotics with six atoms thiazole ring. Sadly, blaCTX-M-1/3/9 genes developed in plasmids of many enterobacteria to degrade cefotaxime and related cephalosporins (1970's) and scientist prepared synthetic carbapenem drugs (1980's) that were very popular now for extensive-drug-resistant pathogens. Soltys discovered in 1952 that M. tuberculosis avian type grown in Dubos medium was more sensitive to penicillin than bovine and human types whereas M. phlei and BCG strains were the least sensitive. Kasik and Peacham in 1968 reported that M. smegmatis (N.C.T.C. 8158), M. fortuitum and M. phlei (MPI) produced a constitutive cellbound β-lactamase that had penicillinase and cephalosporinase activities with different specificities to penicillin-G, ampicillin and early derivative of cephalosporins. Although twenty blaTEM, blaKPC, blaNDM, blaOXA, blaCTX-M like penicillinase isomers were reported in different enterobacteria, only a chromosomal class-A penicillinase (blaC) with homology to blaTOHO and blaTEM genes was reported in MDR Mt. However, few penicillin binding proteins (PBPs) were reported in different Mycobacterium chromosome including Mt [39-43]. Such PBPs have 25%-30% homologies with other bacterial penicillin-binding proteins like E. coli. E. coli possesses three class-A PBPs (PBP1a=OSL52747, PBP1b=AJF05047 and PBP1c=AKM36075) and two class-B PBPs (PBP2/penicillin resistant protein=AAB40835 and PBP3/ peptidoglycan-binding protein=CAA38861) and five Class-C E. coli PBPs (PBP4=CAA42070, PBP5=EDV83509, PBP6=EGI51157, PBP7=OAF94491 and AmpH=ANK05711) [44-46]. M. tuberculosis produces two class A PBPs (ponA1 and ponA2), two class B PBPs (PBPA and PBPB) and a lipoprotein sharing some motifs with the class B PBPs. Six classes C type PBP contained in Mt: one PBP4 which sometime truly AmpH1, one PBP5 (dacB) which truly also AmpH class (see, AEJ49667 and AAK45177 wrongly assigned as PBP4), one PBP6 (DacB1), one PBP7 (DacB2) and the putative type-AmpH which we showed here to be four sub-classes. Our Mt database search pinpointed the misuse nomenclature of Mt PBPs throughout the PubMed and NCBI Database. We have proved that Rv3627c, Rv1730c, Rv1367 and Rv1497 enzymes belong to AmpH class (Rv1730c only described as AmpH and Rv3627c as DacB or Pbp5 in database; AN:AL123456). The AmpH is a bifunctional DDendopeptidase and DD-carboxypeptidase. Which cleaves the cross-linked dimers tetrapentapeptide with efficiencies K2/ k=1,200 M-1S-and removes the terminal D-alanine from muropeptides with a C-terminal D-Ala-D-Ala dipeptide. However, a weak β-lactamase activity reported (1/1,000 the rate obtained for AmpC) for nitrocefin as substrate and the enzyme could be inhibited by 40 μM cefmetazole [47]. It was reported that E. coli AmpH binds to penicillin-G, cefoxitin, cephalosporin-C, cefmenoxime, cefotaxime but poorly with ampicillin, amoxicillin and carbenicillin [48]. The DacB1 (PBP6) and DacB2 (PBP7) related to class-A blaTEM group β-lactamases. The Rv2864c is related to blaOXA-23 and Rv2864c related to blaOXA-58 likely better perform carbapenem hydrolysis. No PBP related to E. coli PBP3 was found in Mt chromosome and PBP4 wrongly described and authentic Mt PBP4 enzymes were found in the database. We pinpointed here that M. tuberculosis AmpH gene was not one in the chromosome but blaAmpH1, blaAmpH2, blaAmpH3 and blaAmpH4 four such class-C β-lactamases would be regulated the degradation of cephalosporins. We also found 17nt sequence from AmpH4 gene had homology to ISKpn6 transposase gene of plasmids indicating new gene creation was mediated by DNA rearrangement involving IS-elements. We also made primers for all Mt PBPs to check their presence and location in the chromosome after WGS. The PubMed search with “AmpH+Mycobacterium” did not produce any result indicating role of AmpH gene in Mt was never investigated carefully.
Materials and Methods
The NCBI GenBank database used to get proteins and nucleotides for PBPs and β-lactamases. The BLASTN used to search nucleotide homology and BLASTP used to search protein homology. Multiple alignment of proteins was done by MultAlign software and multi-alignment was done by CLASTAL-Omega software [49-51]. We found some extended PBPs at the NH2- terminus and likely due to usage of upstream ATG codon instead UTU and UUT initiation codons found in many Mt genes. We found authentic Mt PBP4 in strain FDAARGOS_757 (AN: CP054013/ protein id AUP68265). But our attempts to find Mt PBP3 was failed as BLAST search limited to 5000 sequences only instead search for 20000 sequences were possible few years ago. We used “AmpH+Mycobacterium” to search PubMed but no result indicating role of AmpH β-lactamase in Mt never investigated carefully. We searched 4.4 million bases of Mycobacterium chromosome by naked eye in the computer GenBank database and it took 5 hours. We did BLAST-2 search with DNA to find homology and then homology sequence searched by BLAST-X to get important protein homologues if present in the Mycobacterium chromosome. Once we got the protein isomer, we back compared their penetration in the database by BLAST-P search with increasing sequence parameter to 5000 instead 50. Vibrio species had two chromosomes and we had analysed both separately. The E. coli PBPs and β-lactamase proteins used as standard but sometime K. pneumoniae, S. marcessens, S. aureus, M. leprae, M. intracellulare genomes and proteins were compared.
Results
We used E. coli PBPs protein ids for BLASTP similarity search with 25-30% similarity and greater than 40% cover with Mt PBPs (Supplementary Figures S1-S9). The PonA1 (823aa), PonA2 (810aa), PBPA (491aa), PBPB (605aa), MecA_N (582aa), blaC (307aa), PBP4 (473aa), DacB1 (386aa), dacB2 (291aa), AmpH1 (532aa), AmpH2 (377aa), AmpH3 (428aa) and AmpH4 (517aa) were the PBPs and β-lactamases reported in the database in Mt (Supplementary Figure 2). We found GTG initiation codon for PonA1, PBPB and PBP4 genes whereas TTG initiation codon for AmpH1 and AmpH4 genes (Supplementary Figure 1). The PBP1A and PBP1B were the major transpeptidasestransglycosylases similar to M. tuberculosis PonA1 and PonA2 penicillin-binding-proteins. The PBP2 was assigned as PBPA and PBPB in M. tuberculosis genome. The sequence of PBP3 of E. coli did not match to Mt chromosome but some author put PBPB as PBP3. The DacA, DacB and DacC of E. coli referred as DacB (PBP5), DacB1 (PBP6) and DacB2 (PBP7) in M. tuberculosis. However, we found a MecA_N enzyme was implicated as PBP5 and sometime an AmpH homologue referred as PBP5 in Mt. The Table 1 showed the classification and localization of different PBPs and β-lactamases whereas Figure 1 showed the circular chromosomal localization of those proteins. We first got those enzymes from genomes of M. tuberculosis strain CCDC5180 (AN:CP001642) and M. tuberculosis strain H37Rv (AN: AL123456) (Table 1). Blast search however, could not find an E. coli PBP3 homologue. We did not find any homology between E. coli PBP4 and the published M. tuberculosis PBP4 which we found to be AmpH. Recently, Kumar et al., demonstrated that ponA1 and ponA2 could hydrolyse carbapenem drugs much better than blaC β-lactamase which implicated as sole β-lactamase in Mt. The BLASTP homology search could not find any similarity to bacterial MBL β-lactamases like blaOXA-23, blaOXA-48, blaOXA-51, blaOXA-58, blaKPC-1, blaNDM-1, blaIMP-1, blaGIM-1, blaVIM-1, and blaGES with PBP3 and PBP4 of E. coli. As expected some homology to blaDHA or blaACC with AmpC β-lactamase (protein id. CCP44496) and blaTEM with blaC β- lactamase (protein id. CCP44847) were found. Similarly, we BLASTP homology searched Mt PBP4 (protein id. CCP44126) with many β-lactamases found 25% (60% cover) similarity with class-C β-lactamases (blaAmpC, blaDHA, blaAAC, blaCMY) suggesting such class-C PBP had some capacity to hydrolyse penicillin drugs.
| Name of PBP | CP001642 | Functions | AL123456 |
|---|---|---|---|
| pbpA/Rv0016c (Class B PBP2) | AEJ48854 (482aa), nt. 20117-21565 | Penicillin-binding-protein, FtsI, PbpA, pbp2 | CCP42738 (491aa), nt. 18759-20234 © |
| PonA1/Rv0050c (Class A PBP1A) | AEJ48885 (784aa), nt. 54858-57212 | MrcB, Peptidoglycan transglycosylase- D,D-transpeptidase, Pbp1a | CCP42772 (678aa), nt.53663-55699 |
| Pbp4a/Rv3627c (Class C; DacB) | AEJ49667 (473aa), nt. 1006473-1007894 | Endopeptidase/serine hydrolase, AmpH1 | CCP43655 (532aa), nt. 1010136-1011734; 59aa extension at NH2 |
| Pbp4/Rv1367c AmpC | AEJ50426 (401aa), nt. 1537208-1538413 | β-lactamase, AmpH2 | CCP44126 (377aa), nt. 1539512-1540645 © |
| Rv1497, β-lactamase LipL esterase | AEJ50221 (429aa), nt. 1687057-1688346 | β-lactamase, AmpH3 | CCP44258 (429aa) nt. 1689230-1687944 © |
| blaDHA/Rv1730c (AmpH) | Deletion (no protein), nt. 1945765-1947296 | CP000611/ABQ73489; nt. 1957210-1958763; (Mt H37Ra), AmpH4 | CCP44496 (517aa), nt. 1955692-1957245 © |
| blaC/blaA/Rv2068c (blaTEM) | (no protein), nt. 286834-285911 +/- | CP000611/ABQ73843nt. 2335786-2336709 (Mt H37Ra) blaC | CCP44842 (307aa); nt. 2325886-2326809 © |
| pbpB/Rv2163c (Class B PBP2) | AEJ50811 (605aa), nt. 2417473-2419290 | Fts1 domain, PbpB, D,D- transpeptidase | CCP44940 (679aa), nt. 2425048-2427087 © |
| PbpC/Rv2864c (MecA) | AEJ51432 (582aa), nt. 3156295-3158043 | Fts1 domain, transpeptidase lipoprotein/mec A_N | CCP45666 (603aa) nt. 3175454-3177265 © |
| DacB2/Rv2911c (Class C PBP7) | AEJ51473 (287aa), nt. 3199285-3200160 | DacB2, D,D-carboxypeptidase | CCP45713 (291aa), nt. 3218339-3219214 |
| DacB1/ Rv3330c (Class C PBP6) | AEJ51876 (392aa) nt. 3707205-3708387 | DacB1, D,D-carboxypeptidase | CCP46151 (405aa), nt. 3715777-3716994 |
| Rv3627c Conserved protein | AEJ52151 (450aa), nt. 4059924-4061309 V2S mutation | DD-carboxypeptidase; PBP4 | CCP46450 (461); nt. 4065900-4067284 |
| PonA2/Rv3682 (Class A Pbp1B) | AEJ52200 (810aa), nt. 4116358-4118790 | MrcB, PonA2, D,D- transglycosylase- transpeptidase, Pbp1b | CCP46506 (810aa), nt. 4121916-4124348 |
| Rv3762c β-lactamase? | AEJ52278 (626aa), nt. 4201440-4203320 | Alkyl sulfatase and MBL-hydrolase? | CCP46589 (626aa), nt. 4206996-4208876 © |
Note: Rv0907 located at nt. 1687060-1688346 or NP_215422 (532aa) same to CCP43655 or AmpH. Rv1922 is a lipoprotein (CCP44689; 371aa) and may not be PBP.
Table 1: Localization of penicillin-binding-proteins and beta-lactamases in M. tuberculosis CCDC5180 genome (acc. no. CP001642) and compared with M. tuberculosis H37Rv genome (acc. no. AL123456) genome.
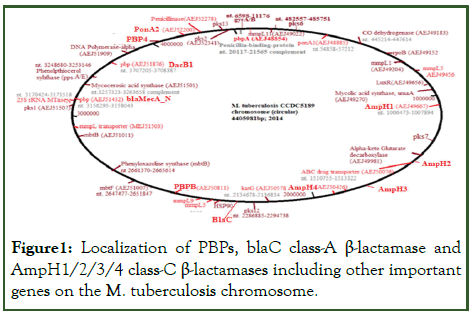
Figure 1: Localization of PBPs, blaC class-A β-lactamase and AmpH1/2/3/4 class-C β-lactamases including other important genes on the M. tuberculosis chromosome.
However, database wrongly described AmpH as PBP4 and sometime DacA. There was no similarities of E. coli PBP5, PBP6, PBP7 to AmpH (Table 2). Whereas, Mt PBPA and PBPB had similarity to blaOXA-23 and blaOXA-58 (26%/14% cover; protein id. AET95879 and 40%/9% cover, protein id. AGC92783) and might show MBL activities hydrolysing carbapenems. Further, Mt Pbp6 and Pbp7 have 27% (37% cover) similarity to Amp (pBR322 plasmid-derived first mdr gene discovered giving resistant to penicillin-G) and blaTEM-1 proteins (protein ids. AAB59737 and AKA86566) but not to class-C blaAmpC or class-D blaNDM-1 or blaOXA-48 suggesting that PBP6/7 will hydrolyse ampicillin but not carbapenem drugs. The PBP3 of E. coli (protein id. CAA38861, AN: X55034, nt. 7943-9709) had no similarity to PBP4 and surprisingly did not detected any PBP3 gene sequence in Mt genomes (AN: CP044345, CO040688, CP089775, CP040691, CP025597, CP054013, CP110619 and CP089927). The disclosed PBP4 gene in the database actually blaAmpH genes and truly PBP4 also found in the database with difficult. We found protein ids AIH68780, CNW39949, WP_003899610, AEJ48551, WP_003902671, AGL29101, NKE28321, CEZ25322 and EGB26878, as true PBP4 of M. tuberculosis with 34% similarity to E. coli PBP4. Such true PBP4 also found in M. bovis (protein id. AHM09405), M. avium (protein id. WP_003873261), M. intracellulare (protein id. AFC46805), M. colombiense (protein id. OBJ06628), M. europaeum (protein id. WP_085241428), M. timonense (protein id. GFG99282), M. gordonae (protein id. OBK44162), and M. liflandii (protein id. VLL08577). Thus, after careful homology analysis, we found four blaAmpH genes regulating the cephalosporin hydrolysis in M. tuberculosis (see below). Interestingly, AmpH1, AmpH2, AmpH3 and AmpH4 full length genes had no similarities to E. coli (AN:LN832404), S. aureus (AN:BX571856), S. pneumoniae (AN:NZ_CP107038) and V. cholerae (Ch-1, AN:NZ_CP028892; Ch-2, AN:NZ_CP012998) chromosomes although proteinprotein homology was detected. Thus, we assume that E. coli has only one AmpH gene that has close similarity to other bacterial species.
| Bacterial Bla gene isomers | GenBank Protein id | PonA1 Rv0050 (PBP1A) CCP42772 | PonA2 Rv3682 (PBP1B) CCP46506 | PbpA Rv0016c (Pbp2) CCP42738 | PbpB Rv2163c (Pbp2) CCP44940 | AmpH2 Rv1367c CCP44126 (Pbp5?) | mecA_N Rv2864c CCP45666 (MBL?) |
|---|---|---|---|---|---|---|---|
| blaTEM-1 | AKA86566 | no | no | 41.3/5 | no | no | no |
| blaSHV-1 | BBW56246 | no | no | no | no | no | no |
| blaCTXM-1 | CAA63262 | no | no | no | no | no | no |
| blaCTXM-2 | ATQ37988 | no | no | 48/5 | no | no | no |
| blaCTXM-9 | AAZ30046 | no | 40/3 | no | no | no | no |
| blaCARB-1 | TPA13973 | no | no | no | no | no | no |
| blaPSE-1 | AKD43563 | no | no | no | no | no | no |
| blaOXA-1 | AAA91586 | no | no | no | no | no | no |
| blaOXA-2 | CAC82805 | no | no | 48.1/5 | no | no | 50/2 |
| blaOXA-23 | AET95879 | no | no | 22.7/22 | 41.3/4 | no | 18.5/34 |
| blaOXA-48 | CDP90224 | no | no | 22.9/8 | no | no | no |
| blaOXA-51 | AQZ59635 | no | no | 38.7/17 | 23.8/15 | no | 25.7/16 |
| blaOXA-58 | AGC92783 | no | no | no | 27.1/11 | no | 26.1/14 |
| blaKPC-1 | AAG13410 | no | no | no | no | no | no |
| blaIMP-1 | AAB30289 | no | no | no | no | no | no |
| blaVIM-1 | CAC82722 | no | no | no | no | no | no |
| blaNDM-1 | AGC54622 | no | no | no | no | no | no |
| blaAmpC | AAD28044 | no | no | no | no | 24.5/60 | no |
| blaACC-1 | CAB46491 | no | no | no | no | 23.1/60 | no |
| blaDHA-1 | ABN58447 | no | no | no | no | 25.5/60 | no |
| blaCMY-1 | CAA63264 | no | no | no | no | 25/61 | no |
| blaGIM-1 | AEX25999 | no | no | no | no | no | no |
| blaToho-1 | BAA07082 | no | no | 48/5 | no | no | no |
| blaGES-1 | AAF27723 | no | no | no | no | no | no |
Table 2: Demonstration of different M. tuberculosis class-A (PonA1 and PonA2) and class-B (PbpA and PbpB) PBPs homologies with different plasmid-mediated β-lactamases.
Then, we organized many Mt low molecular weights class-C PBPs in a table as compared to E. coli class-C enzymes including blaTEM, blaAmpC and blaOXA-58 which was a signature of carbapenemase activity (Table 3). The table stated that few PBPs designated as PBP4 but had no similarity to E. coli PBP4 suggesting wrong nomenclature (red). It was clearly found such enzymes had similarity to AmpH class-C enzymes and such information was wrong many sequences. We continued our search to Mt genomes and we had shown in Table 4 that β-lactamases reported in Mt GG-36-11 (AN:CP025597) and Mt FDAARGOS_757 (AN: CP054013) as compared to Mt H37Rv genome (AN: AL123456): The data is important as we have identified the authentic PBP4 in Mt strain FDAARGOS_757 (protein id. AUP69687) with 34% similarity to E. coli PBP4. BLASTP search identified numerous Mt PBP4 but data still disclosed as conserved protein, unidentified protein or simple β-lactamase-like protein. It was thus clear that Mt PBP4 was wrongly described in many papers and in many genome sequences. Thus, PonA1 and PonA2 (class-A) or PBPA and PBPB (class-B) nomenclatures are very popular and MecA_N (Pbp5), DacB1 (PBP6) and DacB2 (PBP7) nomenclatures are also getting easier to describe Mt PBPs. However, concept of Mt PBP3 is also quite misleading everywhere in the database. In the Table 3 we showed many PBP4 described were truly had no similarity to E. coli PBP4 and they were blaAmpC class enzymes (upper panels). Table 2 suggested PBPA and PBPB had blaOXA-23 activities whereas PonA1 and PonA2 had no similarity to any β-lactamases. Table 3 however, disclosed that DacB1 (PBP6) and DacB2 (PBP7) had blaTEM similarities whereas very important and potent similarities of AmpH1, AmpH2, AmpH3 and AmpH4 enzymes with all class-C β-lactamases like blaAmpC, blaACC, blaDHA as well as blaCMY to lesser extent. It was known that class-C β-lactamases control the hydrolysis of cephalosporin drugs but not carbapenem drugs. The multialignment similarity of those AmpH isomers were shown in Figure 2. It was found that the similarities of AmpH1, AmpH2, AmpH3 and AmpH4 proteins with E. coli AmpH protein obtained as 22.6% similarity/87% cover, 26.55% similarity/49% cover, 28.87% similarity/49% cover and 23.81% similarity/88% cover respectively. Still consistence similarity with blaAmpC were very important for blaAmpH1, blaAmpH2 and blaAmpH4 but the blaAmpH3 had no similarity to blaAmpC but to blaDHA (23.5%/26% cover). Importantly, Mycobacterium AmpH proteins were conserved among species and their AmpC similarities were described in the database. Thus, we are very optimistic about their functions as class-C β-lactamases. Because the BLAST-2 alignment of a plasmid AmpH protein (protein id. WJR74365) from Enterobacter hormaechei shared 23.2%/47%, 26.71%/67%, 30%/10% and 24.61%/79% (similarity/cover) with AmpH1, AmpH2, AmpH3 and AmpH4 enzymes of Mt respectively. But when we did similar search with a Vibrio parahaemolyticus chromosomal AmpH protein (protein id. KKY41027), we found a decent homology 29.95%/49% cover for blaAmpH3 of Mt suggesting we were very right in our analysis of class-C β- lactamases. Similarly, we also BLAST-2 analysed with Yersinia pekkanenii AmpH (protein id. CNI55383) to get good homologies with Mt AmpH proteins as follows: AmpH1=24.05%/56%; AmpH2=27.85%/45%; AmpH3=25.77%/45% and AmpH4=24.13%/71% (%similarity/ % cover) and very similar as compared with E. coli AmpH enzyme (Supplementary Figure 9).
|
Bacterial Bla genes |
Protein id | CCP43655 Rv3627c (AmpH1) | CCP44126 Rv1367c (AmpH2) | CCP44258 Rv1497 blaAmpH3 | CCP44496 Rv1730c (AmpH4) | CCP46151 Rv3330 (DacB1/Pbp6) | CCP45713 Rv2911 (DacB2/Pbp7) | CCP46450 Rv3627c (Pbp4) |
|---|---|---|---|---|---|---|---|---|
|
blaTEM-1 |
AKA86566 | no | no | no | no | 23.2/23 | 26.8/37 | no |
|
blaSHV-1 |
BBW56246 | no | no | no | no | no | 26.4/34 | no |
|
blaCTXM-1 |
CAA63262 | no | no | no | no | no | no | no |
|
blaCTXM-2 |
ATQ37988 | no | no | no | no | 33.9/13 | no | no |
|
blaCTXM-9 |
AAZ30046 | no | no | no | 26/11 | 43.3/7 | no | no |
|
blaCARB-1 |
TPA13973 | no | no | no | 26.6/15 | no | 29.2/27 | no |
|
blaPSE-1 |
AKD43563 | no | no | no | no | 31/29 | 29.3/26 | no |
|
blaOXA-1 |
AAA91586 | no | no | no | no | no | no | no |
|
blaOXA-2 |
CAC82805 | no | no | no | no | no | 52.9/5 | no |
|
blaOXA-23 |
AET95879 | no | no | no | no | no | 42.8/8 | no |
|
blaOXA-48 |
CDP90224 | no | no | no | no | 50/4 | 55/6 | no |
|
blaOXA-51 |
AQZ59635 | no | no | no | no | no | 42.3/8 | no |
|
blaOXA-58 |
AGC92783 | no | no | no | no | no | 40.7/9 | no |
|
blaKPC-1 |
AAG13410 | 26.2/35 | no | no | no | no | 47/5 | no |
|
blaIMP-1 |
AAB30289 | no | no | no | no | no | no | no |
|
blaVIM-1 |
CAC82722 | no | no | no | no | no | no | no |
|
blaNDM-1 |
AGC54622 | no | no | no | no | no | no | no |
|
blaAmpC |
AAD28044 | 22.6/46 | 24.5/60 | no | 28.2/38 | no | no | no |
|
blaACC-1 |
CAB46491 | 29.4/36 | 23.1/60 | 23.2/21 | 26.6/62 | no | no | no |
|
blaDHA-1 |
ABN58447 | 29/46 | 25.5/60 | 23.5/26 | 26/62 | no | no | no |
|
blaCMY-1 |
CAA63264 | 35.5/16 | 25/61 | no | 25.4/46 | no | no | no |
|
blaGIM-1 |
AEX25999 | no | no | no | no | no | no | 46/5 |
|
blaToho-1 |
BAA07082 | no | no | no | no | 33.9/13 | no | no |
|
blaGES-1 |
AAF27723 | no | no | no | no | n0 | no | no |
Table 3: Demonstration of different Class-C β-lactamases in Mycobacterium tuberculosis (% similarity/ % cover); (no means no similarity at all during BLAST-2 search).
| Mt Pbp4 in the database as red colour (aa) | E. coli AmpH, CAD6020925 | E. coli Pbp4, CAA42070 | E. coli Pbp5, DacB,EDV83509 | E. coli Pbp6, DacB1, EGI51157 | E. coli Pbp7, DacB2, OAF94491 | E. coli blaTEM, ALJ52334 | E. coli blaAmpC, AAD28044 | E. coli OXA-58, WEY35876 |
|---|---|---|---|---|---|---|---|---|
| AEJ50426 (418)* Pbp | 23.8/81 | no | no | no | no | no | 28.1/47 | no |
| CCP44126 (377)# Pbp4 | 23.6/58 | no | no | no | no | no | 24.5/60 | no |
| CCP43655 (532) AmpC | 25.5/46 | no | no | no | no | no | 22.6/46 | no |
| CCP44496 (517) Pbp4 | 23.8/65 | no | no | no | no | no | 28.3/38 | no |
| SGJ61490 (488)# Pbp4 | 23.5/69 | no | no | no | no | no | 28.2/40 | no |
| EFD53848 (538) Pbp | 23.8/63 | no | no | no | no | no | 28.3/36 | no |
| AAK45177 (562)# Pbp4 | 22.7/61 | no | no | no | no | 48/4 | 22.6/43 | no |
| CFA99975 (562)# Pbp4 | 22.7/61 | no | no | no | no | 48/4 | 22.6/43 | no |
| KBF69242 (562) AmpC | 22.7/61 | no | no | no | no | 48/4 | 26.6/43 | no |
| KCS24044 (562) AmpC | 22.7/61 | no | no | no | no | 48/4 | 22.6/43 | no |
| CKM67045 (488)# Pbp4 | 24.3/62 | no | no | no | no | no | 28.2/40 | no |
| AEJ46795 (447)* Pbp | 23.8/75 | no | no | no | no | no | 28.2/44 | no |
| SGJ61490 (488)# Pbp4 | 23.5/69 | no | no | no | no | no | 28.2/40 | no |
| CCP45713 (291) DacB2 | no | 64.7/5 | 30.9/96 | 28/93 | 26/82 | 26.8/37 | no | 40.7/9 |
| CMR92158 (352) DacB2 | no | 64.7/5 | 30.9/79 | 28.3/76 | 26/68 | 26.8/30 | no | 40.7/7 |
| VTO96214 (330) DacB | no | 60/6 | 31.4/91 | 30.2/92 | 30.1/77 | 29.2/36 | no | 22.6/26 |
| CMR92158 (352) DacB2 | no | 60/4 | 30.9/79 | 23.6/76 | 26.1/68 | 26.9/30 | no | 40.7/7 |
| AEJ47856 (287) DacB2 | no | 64.7/5 | 30.1/95 | 28.1/94 | 26.1/83 | 26.9/29 | no | 40.7/9 |
| VBA37136 (308) DacB | no | 64.5/7 | 31/93 | 29.4/80 | 28.7/78 | 33.3/31 | no | 25.4/30 |
| AEJ47858 (287) DacB2 | no | 64.7/5 | 31.1/95 | 28.1/94 | 26/83 | 26.8/37 | no | 40.7/9 |
| CCP46151 (405) DacB1 | no | no | 27.1/60 | 27.4/69 | 28.4/50 | 22.3/23 | no | no |
| CCP45666 (603) mecA | no | no | no | no | no | no | no | 26/14 |
| AAK47258 (618) mecA | no | no | no | no | no | no | no | 26/14 |
| AEJ47817 (582) mecA | no | no | no | no | no | no | no | 26/14 |
| ACT24153 (618) mecA | no | no | no | no | no | no | no | 26/14 |
| QKI02648 (415) LipE | 23/22 | no | no | no | no | no | 25/34 | no |
| QKI03168, (409) Bla | 26/41 | no | no | no | no | no | 22/42 | no |
| AUP68265 (307) blaC/blaA/penP | no | no | no | no | no | 46/73 | 29.5/16 | no |
| AUP69687 (461) Pbp4 | no | 34.6/58 | no | no | no | no | no | no |
| AEJ48551 (450)* Hypothetical protein | no | 34.6/60 | no | no | no | no | no | no |
| NKE28321 (421) DacB $ | no | 34.6/64 | no | no | no | no | no | no |
| MBX4339850 (461) DacB$ | no | 34.6/60 | no | no | no | no | no | no |
| CNW39949 (461) | no | 34.6/58 | no | no | no | no | no | no |
| DacB_1 $ | ||||||||
| ULL08577 (461) Pbp | no | 33.3/58 | no | no | no | no | no | no |
| WP_003899610 (461) DacB/Pbp4 | no | 33.6/60 | no | no | no | no | no | no |
Note: Similarity: pbp4/pbp5(0/0); pbp5/pbp6 (60/98); pbp5/pbp7 (26.5/61); pbp6/pbp7(27.6/61); ampH/pbp4 (0/0); pbp4/blaTEM (0/0) * indicates resistant to isoniazid, rifampicin, streptomycin and ethambutol. # wrongly stated as Pbp4 which has no similarity to β-lactamases or 34% similarity to E. coli Pbp4. $ indicated PBP4 wrongly named as DacB.
Table 4: The homology among E. coli different Mt PBPs described as class C penicillin-binding-protein (Pbp4 in majority cases) in the database as compared to E. coli PBPs and β-lactamases (% similarity/ % cover).
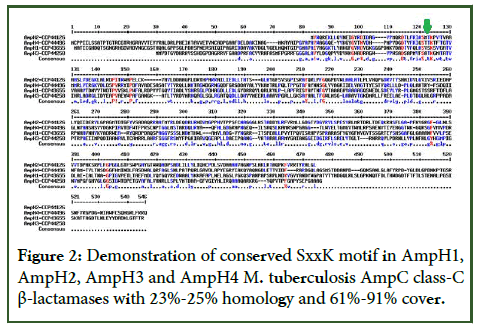
Figure 2: Demonstration of conserved SxxK motif in AmpH1, AmpH2, AmpH3 and AmpH4 M. tuberculosis AmpC class-C β-lactamases with 23%-25% homology and 61%-91% cover.
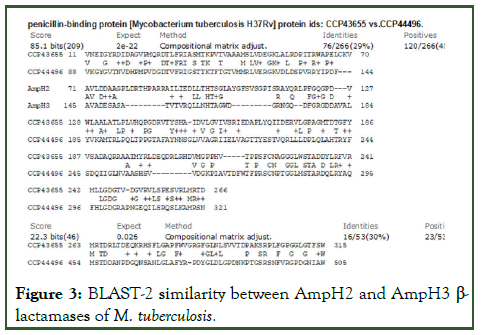
The interesting finding that many AmpH enzymes were described as PBP4 as seen in protein ids. AEJ49667, CNW35153, CKM56251, CMR84911, CMA68255, CKR07362 and COW38226 (Table 3). The serine hydrolases (protein ids. OBA76072, OBA64464, KUP04565) had very strong similarities to AmpH enzymes (protein ids. VBA58402, VBA42298) and almost indistinguishable by BLASTP search. Similarly, lipL esterase enzyme described in the Mt chromosome database also had strong similarity to E. coli AmpH β-lactamase. Combined together, we selected blaAmpH1, blaAmpH2, blaAmpH3 and blaAmpH4 as true class-C β-lactamases of M. tuberculosis controlling hydrolysis of cephalosporin drugs. The BLAST-2 similarities were shown in Figures 3 and 4 demonstrating quite 30% homology between them. Although, multiple alignment reduced such similarity (Figures 5-7).
Figure 3: BLAST-2 similarity between AmpH2 and AmpH3 β-lactamases of M. tuberculosis.
Figure 4: BLAST-2 similarity between AmpH1 and AmpH3 PBPs of M. tuberculosis.
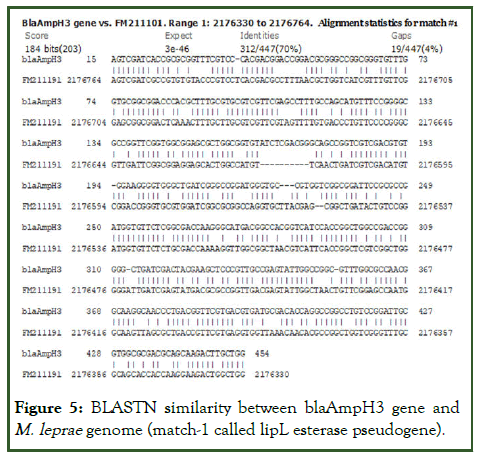
Figure 5: BLASTN similarity between blaAmpH3 gene and M. leprae genome (match-1 called lipL esterase pseudogene).
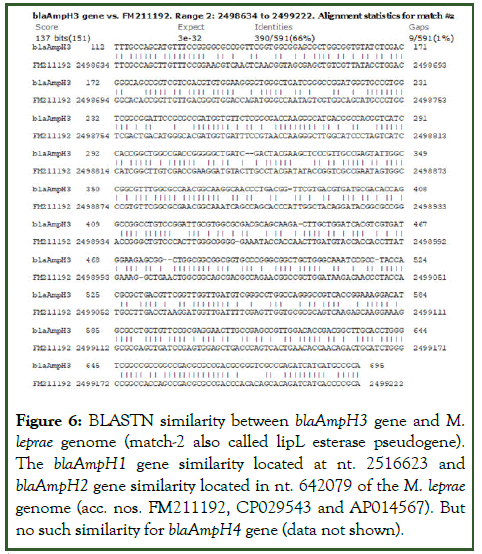
Figure 6: BLASTN similarity between blaAmpH3 gene and M. leprae genome (match-2 also called lipL esterase pseudogene). The blaAmpH1 gene similarity located at nt. 2516623 and blaAmpH2 gene similarity located in nt. 642079 of the M. leprae genome (acc. nos. FM211192, CP029543 and AP014567). But no such similarity for blaAmpH4 gene (data not shown).
Figure 7: Demonstration of blaAmpH3 homologue in M. intracellulare (JAEKMP010000010), M. asiaticum.
(WP_036355751), M. interjectum (LQPD01000066) and other M. species (JACPNU010000009). The data indicated that blaAmpH3 class-c enzymes of Mt has strong similarity to esterase, serine hydrolase and other β-lactamases reported in the database. Actually, such enzymes were AmpH3 class and has less similarity to blaAmpH1, blaAmpH2 and blaAmpH4.
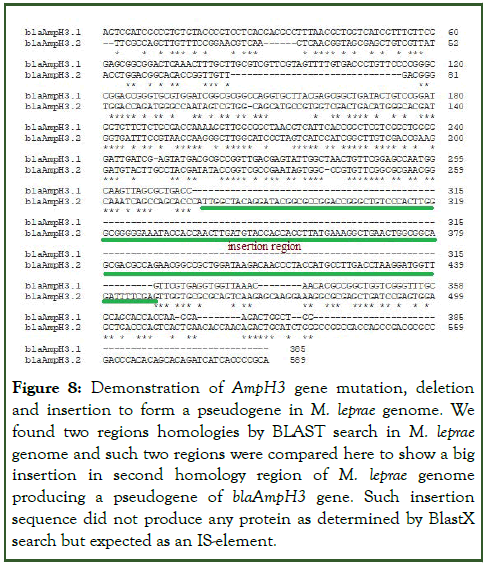
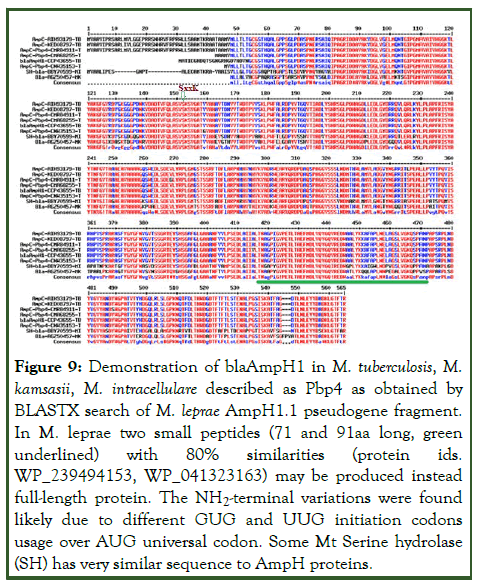
We compared the M. tuberculosis PBPs with M. leprae enzymes (Supplementary Figure 4) because blaAmpH1/2/3 was detected through BLAST-2 search with chromosomes of M. leprae except blaAmpH4. But scanning of WGS were unable to detect the AmpH protein at the chromosomal locus and only found written as pseudogene. The two sequences obtained from homology search with Mt blaAmpH3 gene produced two homology regions. Such two homology sequences when BLAST-2 compared gave homology portions with huge gap demonstrating gene rearrangement (Figure 8). The BLASTX search of AmpH1.1 fragment produced homology peptides but one small peptide with M. leprae and many with other Mycobacterium species including M. tuberculosis. Multi-alignment of those protein clearly indicated such proteins were AmpH3 class as expected but had some mutations among them as in case of an esterase gene or a lipoprotein called lipL gene (Figure 9).
Figure 8: Demonstration of AmpH3 gene mutation, deletion and insertion to form a pseudogene in M. leprae genome. We found two regions homologies by BLAST search in M. leprae genome and such two regions were compared here to show a big insertion in second homology region of M. leprae genome producing a pseudogene of blaAmpH3 gene. Such insertion sequence did not produce any protein as determined by BlastX search but expected as an IS-element.
Figure 9: Demonstration of blaAmpH1 in M. tuberculosis, M. kamsasii, M. intracellulare described as Pbp4 as obtained by BLASTX search of M. leprae AmpH1.1 pseudogene fragment. In M. leprae two small peptides (71 and 91aa long, green underlined) with 80% similarities (protein ids. WP_239494153, WP_041323163) may be produced instead full-length protein. The NH2-terminal variations were found likely due to different GUG and UUG initiation codons usage over AUG universal codon. Some Mt Serine hydrolase (SH) has very similar sequence to AmpH proteins.
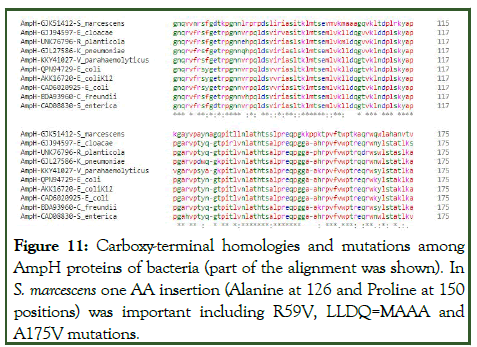
Similarly, repeated experiment with AmpH1.1 and AmpH1.2 fragments of M. leprae produced some homology but no great insertion/deletion was found (one or two nucleotides only). The BLASTX homology detected many protein sequences described as PBP4 of M. tuberculosis but as expected were blaAmpH1 enzymes with some difference at the NH2 terminus initiation point (Figure 10). Interestingly, again we got two small peptide homologies (91 aa and 81 aa long) in M. leprae pinpointing the pseudogene concept. However, we detected PonA1 (protein id. CAC32220), PonA2 (protein id. CAC31824), PBPA (protein id. AWV47033), PBPB (protein id. AWV47654), MecA_N (protein id. AWV48102), DacC (protein id. AWV47526), DacB1 (protein id. AWV47188) all PBPs except blaAmpH1/2/3/4 enzymes in M. leprae. Rationally, M. tuberculosis genome is 4.46 million bases whereas M. leprae has only 3.25 million bases. We speculated that deletions and insertions including point mutations were occurred at the AmpH locus of M. leprae [52]. Thus, we explained the AmpH pseudogenes concept but needed more careful analysis. Never-the-less, we beautifully disclosed the present concept of PBP4 which in most cases were AmpH genes of Mt and true PBP4 was never analysed in Mt. The AmpH gene of many bacteria shared close similarities as demonstrated in Figure 11. But many mutations in the V. parahaemolyticus chromosomal AmpH protein (protein id. KKY4107), Enterobacter hormaechei plasmid-mediated AmpH protein (protein id. WJR74365) and Raoviltella planticola AmpH protein (protein id. UNK76796) were found (Table 5).
| Name of the PBP in Mt GG-36-11 | Protein id for Mt GG-36-11 | AL123456 homology | Name of the PBP in FDAARGOS_757 | Protein id for FDAARGOS_757 | AL123456 homology % |
|---|---|---|---|---|---|
| PbpA | AUP66337 | CCP42738 (100/100) Pbp2a | DacB1 carboxypeptidase | QKI02248 | CCP46151 (100/100) DacB1 |
| PBP1A | AUP66368 | CCP42772 (99.7/82) PonA1 | PonA2 Transglycosylase/ D,D-transpeptidase | QKI02574 | CCP46506 (100/100) PonA2 (PBP1B) |
| AmpC | AUP70055 | CCP43655 (98.7/100) AmpH1? | D-Alanyl-D-Ala- Carboxypeptidase, DacB | QKI02525 | CCP46450 (100/100) Pbp4 |
| β-lactamase | AUP67611 | CCP44126 (100/100) AmpH2 | Beta-lactamase family protein (lipE) | QKI02648 (lipase, LipE) New-1 | CCP43655 (24.2/57) |
| β-lactamase | AUP67734 | CCP44258 (100/100) AmpH3 | PbpA D,D-transpeptidase | QKI02805 | CCP42738 (100/100) Pbp2 (Fts1) |
| PBP | AUP67942 | CCP44496 (100/100) AmpH4 | PonA1 Transglycosylase-DD-transpeptidase | QKI02837 | CCP42772 (99.5/100) PonA1(PBP1A) |
| blaA/blaC | AUP68265 | CCP44842 (100/100) blaC | Beta-lactamase family protein | QKI03645 (Serine hydrolase) | CCP43655 (99.6/96) AmpH1? |
| PbpB | AUP68350 | CCP44940 (100/100) Pbp2b | Beta-lactamase family protein | QKI04082 | CCP44126 (100/100) AmpH2 |
| PBP | AUP68992 | CCP45666 (100/100) MecA_N | Beta-lactamase | QKI04208 | CCP44258 (100/100) AmpH3 |
| D-alanyl-D-alanine carboxypeptidase | AUP69035 | CCP45713 (100/100) DacB2 | Beta-lactamase family protein | QKI04417 | CCP44496 (100/100) AmpH4 |
| D-alanyl-D-alanine carboxypeptidase | AUP69421 | CCP46151 (100/100) DacB1 | blaA/penP | QKI04734 | CCP44842 (100/100) blaC |
| D,D-endopeptidase | AUP69687 | CCP46450 (100/100)PBP4 | PbpB D,D-transpeptiudase | QKI04820 | CCP44940 (100/100) PbpB |
| Membrane carboxypeptidase | AUP69732 | CCP46506 (100/1000) ponA2 | Beta-lactamase | QKI05481 | CCP45666 (100/100) MecA_N |
Note: AUP66689 homologues: AIH44463, AGL25868, KBI52351, AFY28928
Table 5: Demonstration of PBPs and beta-lactamases reported in Mt GG-36-11 (AN: CP025597) and Mt FDAARGOS_757 (AN: CP054013) as compared to Mt H37Rv genome (AN: AL123456): This data is important as identified authentic PBP4 in Mt strain FDAARGOS_757 (+1 site in the chromosomes vary).
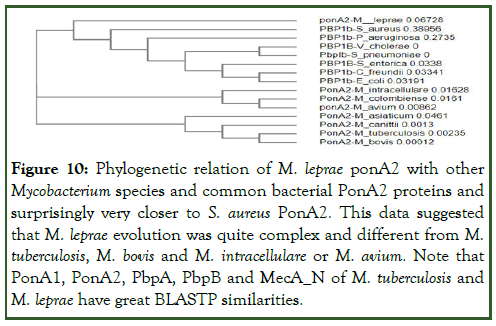
Figure 10: Phylogenetic relation of M. leprae ponA2 with other Mycobacterium species and common bacterial PonA2 proteins and surprisingly very closer to S. aureus PonA2. This data suggested that M. leprae evolution was quite complex and different from M. tuberculosis, M. bovis and M. intracellulare or M. avium. Note that PonA1, PonA2, PbpA, PbpB and MecA_N of M. tuberculosis and M. leprae have great BLASTP similarities.
Figure 11: Carboxy-terminal homologies and mutations among AmpH proteins of bacteria (part of the alignment was shown). In S. marcescens one AA insertion (Alanine at 126 and Proline at 150 positions) was important including R59V, LLDQ=MAAA and A175V mutations.
Truly, the database also pointed penicillin-binding-protein 4, has AmpC motif which was not possible pinpointing wrong nomenclature (Table 4, upper panels). In Table 4 we also shown few PBP4 which appeared as AmpC class enzymes with similarities to blaAmpC, blaDHA, blaACC and blaCMY that all could be hydrolyse the cephalosporins like cefotaxime which widely used now to clear infections. We have not found good similarity to MBLs and carbapenemases (blaNDM-1, blaVIM-1, blaIMP-1) with M. tuberculosis PBPs (Table 2). Whereas PonA1 and PonA2 disclosed as strong hydrolysing activities for carbapenem drugs and we opposed such concept. Truly, we found some similarities of PBPA and PBPB (PBP2 class) with blaOXA-23 or we also found some similarity of blaMecA_N with blaOXA-58 enzyme and that way such enzyme might perform carbapenem binding and hydrolysis. In fact, blaC β-lactamase which also referred as blaA in some cases hydrolysed the penicillin drugs better whereas DacA1 (PBP6) and DacB2 (PBP7) might be considered for such activities as demonstrated by homology search with 24 plasmid-mediated β-lactamases of E. coli and K. pneumoniae (Table 4). The blaC enzyme of M. tuberculosis had similarity to blaZ of S. aureus and blaCARB of V. parahaemolyticus suggesting an important enzyme for penicillin hydrolysis. However, blaC also has similarities to blaCTXM1/ 2/9 considering its activity as cephalosporinase (ESBL). Chromosomal enzymes were found inducible as in blaZ β-lactamase of S. aureus and AmpC enzyme of S. marcescens and P. aeruginosa. However, such induction was lost (blaR1 or ampR) in case such β-lactamase appeared in plasmid and likely multi-copies plasmids increased the gene dosage. Sadly, we are unable to show any induction system in M. tuberculosis and much careful attention may be necessary.
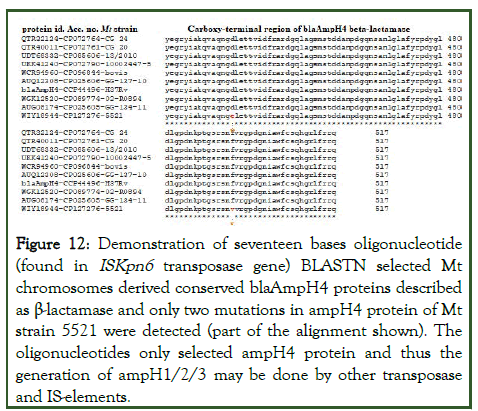
Interestingly, in the Mt strain FDAARGOS_757, the AmpH1 gene SNP was found where one extra-G at nt70 of the AmpH1 gene creating a stop codon TGA at codon 45. However, from the upstream ATG codon the protein was produced with alter 22 AAs at the NH2-terminal. Astonishingly, we found same case in Mt strain GG-36-11 AmpH1 gene and it could a sequencing error. To clarify the issue, we BLASTN search 60nt with or without extra-G to see the penetration of the mutant allele. It appeared normal (Mt strain H37Rv) had only 46 sequences whereas rest thousand were with extra-G. However, such discrepancy did not alter the β-lactamase motif. Now question may appear if more AmpC-like genes in M. tuberculosis? Surely, we found a serine hydrolase protein in Mt strain FDAARGOS_757 with low similarity to AmpH1 (23%/42% cover) with protein id. QKI03168 implicated as β-lactamase as such domains spread to different esterase and lipoprotein and akyltransferases. More surprisingly, large plasmids (accession numbers: NC_012692, LN850163, LC055503, CP007558) with β-lactamases (blaTEM, blaCTXM, blaAmpC, blaSHV) did not show homology with blaAmpH1/2/3/4 genes but only 17nt homology (173580-5’-CCTTGGTGCCGTCGACC-3’-173564) detected in the transposase (protein id. AHY14824) of C. frundii plasmid pKEC-a3c (AN:CP007558) with the blaAmpH4 gene nt. 1236-1252 of M. tuberculosis. Thus, all blaAmpH1/2/3/4 genes involve with higher rearrangement and mutations involving ISelements. Interestingly, BLASTN search with this 17nt sequence selected many plasmids and chromosomes including many M. tuberculosis chromosomes. The AmpC or AmpH genes were chromosomal origin and their origin in plasmids were further checked. Analysis found two K. pneumoniae plasmids (accession nos. CP067912 and AP024793) had homologies to 17nt sequence in the IS1182-like ISKpn6 transposase gene. Similarly, C. freundii plasmids (accession nos. CP054297 and CP110778) hybridised to 17nt sequence in the same IS1182-like ISKpn6 transposase gene. Such finding used to be crucial as we proposed that blaAmpH4 gene was produced by insertion of ISKpn6 transposase although BLASTP search produced no homology between blaAmpH4 protein and ISKpn6 transposase protein. The 17nt sequence however, detected blaAmpH4 sequence in the many chromosomes. The Mt strain 5521 (AN:CP127276, nt. 1946009-1945993) and Mt strain 02-R0894 (AN:CP089774, nt. 195118-1951102) selected by 17nt BLASTN search and in that locus blaAmpH4 protein sequence (protein ids.WJH80555 and WIY18944) were found. However, we also selected a serine hydrolase with protein id. CKR72489, implicated as putative β-lactamase of M. tuberculosis. Thus, we confirmed the role of ISKpn6 sequence and a transposase in the genesis of M. tuberculosis conserved blaAmpH4 gene. Many β-lactamase proteins were selected and multi-alignment was done in presence of blaAmpH4 protein. The data presented in Figure 12 that such β-lactamases were perfectly blaAmpH4 protein and only two AAs mutations (D435E and F495V) were found in the Mt strain 5521 (protein id. WIY18944) at the carboxy-terminus. We also detected a β-lactamase pseudogene in Mt FDAAGROS_756 that was occurred due to deletions of 19nt (5’- AACGGTCATCGACTTCCG-3’) at nucleotide 1313 and a 3nt (5’-CTT-3’) at nucleotide 1481 of AmpH4 gene (Supplementary Figure 5). It was found in Mt strains 5005, TCDC11, H54, WC078, TBV4768, MDRMA1565, LN55, HN-506, SCAID 252.0, SCAID 320.0, PR10, SGF0472017, BT1, 1-0009P6C4, 1-0116P6C4, 2-0068P6C4, SEA02010036P6C4 and CCDC5180 had similar genetic loci. Thus, roles of IS-elements (ISKpn6 transposase) in the creation of AmpH4 gene was postulated and similar events of deletions, mutations and rearrangements could be possible due to over dose use of penicillin, cephalosporin and carbapenem antibiotics.
Figure 12: Demonstration of seventeen bases oligonucleotide (found in ISKpn6 transposase gene) BLASTN selected Mt chromosomes derived conserved blaAmpH4 proteins described as β-lactamase and only two mutations in ampH4 protein of Mt strain 5521 were detected (part of the alignment shown). The oligonucleotides only selected ampH4 protein and thus the generation of ampH1/2/3 may be done by other transposase and IS-elements.
We have decided to make PCR primers specific for all Mt PBPs for RT-PCR to detect their expression and also to check the individual enzyme localization after complete genome sequencing. Such attempt will surely avoid the case of nomenclature problem. The PonA1F primer is 5’-GTA CCA ACC AGG TCT CCA CG-3’ and PonA1R primer is 5’-TCG AGC AAC TCC CTT GTC AC-3’ with PCR product 718bp and Tm of the primers is 60OC. The PonA2F primer is 5’-CGA TTC CAG GCC AAA GGT CT-3’ and PonA2R primer is 5’-TCG ATG AGC TGG TCG ATT GG-3’ with PCR product 404bp and Tm 60OC. The PBPA primers were made and PbpAF=5’- CTG CGG GTC TAT CCC AAT CC-3’ and PbpAR=5’-CCG CCG TAG TTC TCT AGC TG-3’ with Tm=60°C and PCR product size=566bp. Similarly, the PBPB primers were made and PbpBF= 5’-CAC GCT GAT GCT TTC CCA AC-3’ and PbpBR=5’-CCG GCG AAG GTG ATC CAA TA-3’ with Tm=60°C and PCR product size=516bp. The Pbp4F primers is 5’-AAC GCC TCC GAC AAT GTG AT-3’ and Pbp4R is 5’-AAC TTA GTG GCC AGA GCG TC-3’ with PCR product size 485bp and Tm for primers about 60°C. Both primers selected only single 100% match on the Mt genome: As for example, Mt H37Rv-1 genome selected nt. 4072802-4072821(+/-) for forward primer and nt. 4072337-4072356 (+/+) for reverse primer (AN: CP110619) during BLASTN search of oligonucleotides separately.
The eight primers of blaAmpH1/2/3/4 isomers were also made and their localization in the Mycobacterium also confirmed and a restriction enzyme analysis scheme outlined (Tables 6 and 7). It was found that restriction enzymes NotI (383, 173bp), NarI (349, 239bp), MscI (364, 181bp) and NdeI (307, 283bp) cut single in the AmpH1.1, ampH2.1, ampH3.1 and ampH4.1 PCR fragments respectively allowing to differentiate the PCR fragments without DNA sequencing. The 1.2% agarose gel electrophoresis would be required to see the distinct low molecular weight bands in presence of ethidium bromide. Although the primers showed 100% homology with Mt H37Rv-1 and quite high similarities with M. bovis but M. marinum, M. kansasii, M. avium, M. leprae and M. intracellulare did not showed 100% homology. Thus, all primers could be assigned as Mt specific but high annealing temperature (54°C) should be maintained during PCR to reduce unwanted PCR bands.
| Primer name | Sequence of the primers | Tm (°C) | Size, bp | Res cut (bp) | Res cut (bp) |
|---|---|---|---|---|---|
| blaAmpH1F | 5’-GACCTCTACTCGCATCGCTC-3’ | 60.04 | 556 | SacII=556 | NotI=383, 173 |
| blaAmpH1R | 5’-TTACCCCCACGTTGAATCCG-3’ | 60.04 | |||
| blaAmpH2F | 5’-GCACCGCTGTACCAGATCAT-3’ | 60.18 | 588 | SacII=588 | NarI=349, 239 |
| blaAmpH2R | 5’-TTTCGCCAGGGATGGGTTAC-3’ | 60.03 | |||
| blaAmpH3F | 5’-CTGACGGTTCGTGACGTGAT-3’ | 60.39 | 545 | SacII=461, 84 | MscI=364, 181 |
| blaAmpH3R | 5’-AACCGTATGCCGTCGATCTC-3’ | 59.97 | |||
| blaAmpH4F | 5’-GTGAGCAGATTCTGAGCCGA-3’ | 59.83 | 545 | SacII=427, 118 | NdeI=307, 238 |
| blaAmpH4R | 5’-CTTGTTGTCGGGTCCAAGGT-3’ | 60.18 |
Table 6: Primers for four BlaAmpH isomers and restriction enzyme analysis.
| Primers name | M. tuberculosis CP110619 | M. bovis AP010918 | M. avium CP033911 | M. leprae AL450380 | M. kansasii CP006835 | M. intracellulare CP012885 |
|---|---|---|---|---|---|---|
| blaAmpH1F | 1010638 | 1012826 | no | no | 2108763 | no |
| blaAmpH1R | 1011193 | 1013381 | no | no | no | no |
| blaAmpH2F | 1540162 | 1539557 A=G at 16* | no (8-19) | no (1-12) | no (3-14) | no (4-18) |
| blaAmpH2R | 1539575 | 1538970 | no (3-14) | no (3-15) | no (1-13) | no (2-14) |
| blaAmpH3F | 1688535 | 1686288 | no (5-17) | no (2-15) | no (3-16) | no (3-16) |
| blaAmpH3R | 1689079 | 1686832 | no (8-20) | no (7-19) | no (7-20) | no (8-20) |
| blaAmpH4F | 1956543 | 1934390 | no (3-14) | no (9-20) | no (1-12) | no (3-20)# |
| blaAmpH4R | 1955999 | 1933846 | no (7-20) | no (8-19) | 1665625 A=G at 16* | no (7-19) |
Note: The position in the genome with 100% similarity shown. In the bracket () homology region in the 20nt primers were given. * indicated one nucleotide mismatch only. # indicated quite high homology.
Table 7: Homology of AmpH1/2/3/4 primers with different Mycobacterium species.
The primers for mecA_N gene (protein id. CCP45666) were also made and mecAF=5’-CAT CCC AAG CTA GGC GAA CA-3’ and mecAR=5’-CAA CGT GAT CGA AAC CGA CG-3’ with Tm 60°C and the product size=522bp. The mecA_N has similarity to blaOXA-58 (19%/58% cover) and also BLASTP search identified conserved motifs with blaOXA-23. We designated this PBP as PbpM and sometime referred as Pbp5 or DacB but has no similarity to E. coli PBP5 or DacB. The primers for blaC (sometime referred as blaA) were made and blaCF= 5’- GCT CAC GCA TCT GGA CAA AC-3’ and blaCR=5’-CCA TCC AAT CGG TGA GCA GT-3’ with Tm=60°C and the product size=454bp. The primers of DacB1 was also made and DacB1F=5’-ACG CTC ACC CTC AAC AAG TC-3’ and DacB1R=5’-GTT GAA CCC GTA GTC GAG CA-3’ and Tm=60°C and the product size=594bp. The primers for DacB2 were also made and dacB2F=5’-CCA AGA TGA ACG CCA AAG CC-33’ and DacB2R=5’-TCG CAG CCT GAT CCC AAT AC-3’ and the Tm=60°C and the product size=390bp. Thus, after complete sequencing of genome of Mt, the position of individual PBP could be assigned using any forward for reverse primers and BLAST-2 search.
Discussion
Escherichia coli possesses five high molecular weight PBPs, with three (PBP1a, PBP1b, and PBP1c) in class-A and two (PBP2 and PBP3) in class-B. This organism has seven low-molecular-weight class-C PBPs: D,D-carboxypeptidases or D,D-endopeptidases (PBP4, PBP5, PBP6, PBP6b, PBP7, PBP4b, and AmpH). Using BLASTP and BLASTN homology searches, we described the correct nomenclature of Mt PBPs. Database analysis suggested a wide distribution of mutations among the different β-lactamases. Mutations affect the specificities to β-lactams hydrolysis. As for example, mutation of 238S or 240K of blaSHV β-lactamase increased specificities to ceftazidime and cefotaxime respectively as comparable to blaCTXM-9 which could hydrolysis cephalosporin drug efficiently. The ESBLs blaTEM-155 (Q39K, R164S, E240K) and blaSHV-105 (I8F, R43S, G156D, G238S, E240K) and blaSHV-48 (V119I) mutations affected drug specificities [53]. The sequence similarity between blaTEM-1 and blaNDM-1 was not found but both hydrolysed the penicillin drugs. The blaOXA-1 favours the hydrolysis of oxacillin than ampicillin but blaOXA-23 and blaOXA-51 prefer to hydrolysis carbapenem drugs. Thus, roles of PBPs in penicillin-binding and penicillin-hydrolysis of few hundred penicillin, cephalosporin and carbapenem drugs have a complex mechanism. The original blaCTX-M and blaGES enzymes were ESBLs that hydrolyse both penicillin and cephalosporin well, but not aztreonam and carbapenem drugs and were inhibited by the newer β-lactamase inhibitors such as avibactam, vaborbactam and relebactam. The AmpC β-lactamases are cephalosporinases encoded on the chromosomes of many of the Enterobacteriaceae where they mediate resistance to early cephalosporins like cephalothin, cefazolin, cefoxitin, ampicillin, amoxicillin, and few β-lactamase inhibitor-β-lactam like amoxicillin-cavulinate combinations [54]. We pinpointed here that M. tuberculosis penicillin-resistance was mediated by three chromosomal AmpC-like β-lactamases (blaAmpH1/2/4 whereas other authors neglected such enzymes in their analysis and accepted only one AmpH enzyme in Mt. The analysis corrected that blaAmpH3 is more like blaDHA than blaAmpC. Kumar et al. recently described PonA1/A2 enzymes as best β-lactams binding and hydrolysis although data presented the opposite fact (PonA1/A2=<200 M-1S-1 vs. and AmpH=>4000 M-1S-1). We detected conserved SxxK motif in PonA1/A2, PBPA/B and also AmpH1, AmpH2, AmpH3 and AmpH4 of Mt as well as E. coli blaAmpC and P. aeruginosa blaDHA β-lactamases. Our interpretation was that the major β-lactamase activities in Mt was regulated by AmpH β-lactamases which had similarity to blaAmpC, blaDHA, blaACC and blaCMY. Surely, AmpH1 and AmpH4 will have more class-C β-lactamase character with wider homologies whereas functions of AmpH2 and AmpH3 will be confirmed after laboratory testing with purified enzymes. Our guess that the hydrolysis by PonA1 and PonA2 were happened due to over-expressed enzyme and in vivo effects of those proteins for penicillin drug resistance was not surely happening!
Further, no Mt PBP with homology to E. coli Pbp3 was found and some author described Mt PbpB as Pbp3 in the literature [55,56]. Similarly, AmpH or DacA, DacB1 described as PBP4 in Mycobacterium database. But correct PBP4 enzyme was located in the genomes of Mt strains OA115DS, J09400698, CCDC5079 and ATCC-35743 whereas in standard strain H37Rv reported as conserved protein (AN:AL123456) [57,58]. The carbapenemase homology detected in blaOXA-23-like PBPA/B enzymes (protein ids. CCP42738 and CCP44940) and blaOXA-58 related blaMec_N enzyme (protein id. CCP45666) whereas MBLs like blaVIM, blaIMP and blaNDM homologies could not be found.
The ampE, ampD and ampR virulence genes specifically in Pseudomonas aeruginosa suggested as the regulators of induction and de-repression of AmpC genes. The ampR located upstream of AmpC gene and was a LysR-type regulator. Interestingly we located few LysR-like enzymes in Mt chromosome, upstream of AmpH1, AmpH2, AmpH3 and AmpH4 genes (data not shown). Plasmidmediated AmpC gene was also found recently and blaZ of S. aureus has 30% homology to blaC β-lactamase of Mt and blaCARB gene of V. parahaemolyticus also had BlaC similarity (data not shown). As comparison, an induced blaAmpC gave MIC90 250 μg/ml for cefotaxime but it reduced to MIC90 0.06 μg/ml without such induction [59]. Interestingly BLASTN search with Mt genome did not match greatly with an inducer (ampR) but three loci were detected with GC-rich minor similarities. But we did not find any ampR protein at the upstream of AmpH1, AmpH2, AmpH3 and AmpH4 genes in Mt. The Mt genome had numerous TFs genes like LuxR, ArcR, GntR, LytR, TetR and FadR and thus activation may possible in trans. Thus, our point of view was that AmpH genes were expressed in low amount as compared to inducible AmpC gene found in P. aeruginosa or plasmid-mediated AmpH genes.
Bhakta and Basu purified a truncated PonA2 with huge binding to amoxicillin and cefotaxime (K2/K=50000 M-1S-1) but no comparable data found with Mt other PBPs. Nadler JP et al. described the some PBP-like similarities in Mt but disclosed MT0202 (AAK44422; AE000516.2, nt. 223676-224776), MT0501 (AAK44724; AE000516.2, nt. 573158-574513) and MT1477 (AAK45741; AE000516.2, nt. 1611290-1612105) had no L,D-transpeptidase activities to make 3->3 linkage in Mt instead 4- >3 linkage of peptidoglycan cross linking but MT0125 (AAK44348; AE000516.2; nt.140258-140890) did well. However, such enzymes had no similarities to β-lactamases [60,61]. Thus, D,D-carboxypeptidase PBPs are different enzyme and have found good penicillin-binding. The β-lactams hydrolysis by concentrated enzyme (over-expressed and purified) in vitro may not be comparable to its in vivo function.
Streptococcus pneumoniae DacB acts as an L,D-carboxypeptidase towards the tetrapeptide L-Ala-D-iGln-L-Lys-D-Ala of the peptidoglycan stem, with Km and kcat values of 2.84 mM and 91.49 S-1 respectively [62]. Satishkumar et al., demonstrated that PBP4 of Staphylococcus aureus could not be considered to be a classical mediator of β-lactam resistance and our data analysis supported the similar view in Mt [63,64]. However, S. aureus MecA and PBP4 both had crucial role in penicillin-drug resistance. Recently, an inhibitor of PBP4 was discovered to act as antibacterial drug [65]. Interestingly, data analysis found that S. aureus PBP4 (protein id. ANW82090) had no similarity to E. coli (protein id. CAA42070) or M. tuberculosis (protein id. CCP46450) PBP4 but shared a homology to DacB1 (Pbp6) of E. coli (30.3%/53% cover) and M. tuberculosis (19.92%/53% cover). Thus, correct nomenclature would be S. aureus PBP6 (based on E. coli homology) with blaCTX-M homology (40.7%/96% cover; protein ids. CAA63262, ATQ37988, AAZ30046) although it had no similarity to blaC of M. tuberculosis or blaTEM of E. coli. Thus, the compound 9314848 was an inhibitor of cephalosporinases, a MRSA S. aureus characteristic. Surprisingly, there was report that mutations in PBP4 (V223I and A617T) caused decreased susceptibility to penicillin and carbapenem in Enterococcus faecalis. A penicillin-binding-protein-2 thioesterase was shown to catalyse head-to-tail macrolactamization of non-ribosomal peptides whereas a similar enzyme, SurE had higher chains length peptides specificity [66]. Surely, PBPs have roles in the assembly of cell wall peptidoglycan and D-Ala-D-Ala similarity reflects a penicillinbinding motif. Interestingly, in Mycobacterium PBP2 had two isomers (PBPA and PBPB) to generate early cephalosporin resistance but in Neisseria gonorrhoeae five such isomers were detected [67]. Although data analysis suggested that all PBP2 isomers (protein ids. AAA25463, BAI66126/7/8 and BAB86942) were related to PBPB of Mt. Thus, identification of four blaAmpH isomers regulating in Mt was not surprising. The PonA1/A2 conserved SxxN and SxN penicillin-binding motifs were shown at the carboxy-terminus but KxG conserved motif in PonA1 was changed to “PMS” in PonA2. The conserved SxxK, SxN and KxG amino acid sequences in PBPA, PBPB and PBPC (MecA_N) of M. tuberculosis also demonstrated as compared with E. coli and S. aureus PBP2 (Supplementary Figure 5). The conserved SxxK sequence in AmpH1, AmpH2, AmpH3 and AmpH4 M. tuberculosis β-lactamases were found but no KxG and SxN motifs were detected. As there were no much conserved domain among four AmpH proteins, we predicted extensive small deletions and mutation during selection of those conserved AmpC and DHA β-lactamase motifs. Thus, even we did not find any similarity between ponA1/A2 with any 24 β-lactamases, still some PBP-motifs indeed was present and widely β-lactam hydrolysis were detected by different workers. Interestingly, Wivagg et al., demonstrated that some mutations in the PonA2 gene or inactivation of the gene gave penicillin drug resistance instead penicillin sensitivity demonstrating penicillin-binding and penicillin-cleavage were two different functions [68,69]. The PBP-motif located however, at the carboxy-terminus of the proteins and likely glycosyltransferase activity located at the amino-terminus of PonA1 and PonA2. Similarly, Kieser et al., demonstrated using similar transposon-tagged mutagenesis approach that peptidoglycan synthesizing enzymes like PonA1 plus PonA2 (peptidoglycan transglycosylase and 4,3 transpeptidase) and IdtB (3,3 transpeptidase) proteins have Mycobacterium specific mechanism to make peptidoglycan and binding of penicillin varied considerably among isomers [70]. The Neisseria gonorrhoeae PonA1 gene mutation was also shown to cause penicillin drug resistance [71]. The V. prarahaemolyticus carbenicillin-hydrolysing activity was associated with chromosomal blaCARB gene but PonA1 gene was also located. Dekhil and Mardassi showed recently the important of PonA1, Rv0197, esxK and eccC2 genes mutations in Mt sublineages L4.3 evolution [72]. Thus, PonA1/A2 have roles other than penicillin-binding but carbapenem hydrolysis is debated.
Interestingly, we detected a relation of ISKpn6 transposase and blaAmpH4 gene as 17nt sequence perfectly matched between two un-related genes. We BLASTN searched Mt chromosomes with 17nt oligonucleotides and selected many Mt chromosomes at the blaAmpH4 locus whereas all AmpH4 proteins appeared conserved and only two mutations detected for Mt strain 5521 AmpH4 protein. On the other hand, many bacteria had the one blaAmpH gene and had high similarities although mutations were detected in different Genus of bacteria. Surely, we suggested a role of IS1182-like ISKpn6 transpose in the genome rearrangement of Mycobacterium and such deletions, mutations and rearrangements could cause new gene and also could inactivate another gene as happened in Mt FDAARGOS_756 AmpH4 gene (data not shown).
MβL enzymes alone occupy class B which is subdivided into B1, B2 and B3 sub-classes evolution events and require bivalent metal ions such as Zn2+ and Mg2+ for their activity. The catalytic site of MBL-enzymes contained a conserved motif (HxHxDH) and more downstream residues H196 and H263. In bacteria, more than 325 variants grouped into 63 MβL types and divided into three sub-groups: subgroup B1 (e.g., VIM-2, NDM-1 and IMP-1); B2 (e.g., CphA1 and ImiS) and B3 (e.g., LRA-1, GOB-13 and CAR-1). But such conserved motifs were lacking in PBPs of Mt [73]. Truly, MBL mechanism in Mt was still elusive. The AmpC is originally detected in chromosome (E. cloacae, S. marcescens and P. aeruginosa) but now mobilised into variety of plasmids. The AmpR gene regulates AmpC genes negatively. Certain β-lactams induce the production of cell-wall degradation products (eg, N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid oligopeptides) which compete with uridine diphosphate (UDP)–N-acetylmuramic acid peptides for binding to AmpR. With decreased UDP-N-acetylmuramic acid peptide binding to AmpR, results in higher production of AmpC enzymes [74]. Further ampD and ampG regulates such membrane oligopeptides production and transport. The E. coli class-C β-lactamases like blaCMY, blaFOX, blaDHA, blaACC, blaACT, blaMIR, blaDHA, and blaMOX, produced in plasmids leading to resistance to expanded-spectrum cephalosporins and we found class-C β-lactamases (AmpH1/2/3/4) in Mt.
The DacD gene was cloned which was also known as PBP6B and was located in E. coli (AN: LN832404), K. pneumoniae (AN: NZ_CP101563) and S. enterica (AN: NZ_CP129206) genomes but such gene was not detected in M. tuberculosis (AN: AL123456), M. leprae (AN: AL450380), S. aureus (AN; BX571856), S. pneumoniae (AN: NZ_CP107038) and C. diphtheriae (AN: NZ_CP040523) genomes and also little homology detected with both V. cholerae chromosomes (AN: NZ_CP028892 for Ch-1 and NZ_CP012998 for Ch-2). Thus, we greatly compared the presence of all PBPs in M. tuberculosis. We disclosed four class-C β-lactamases in the hydrolysis of cephalosporin drugs which never described well although presence of AmpH and AmpC class enzyme activity indirectly reported in the NCBI Mt sequence database. The M. leprae was a different entity and caused leprosy in India and many other parts of Asia. M. leprae has genetic instability [75-77]. We analysed in details the localization and characterization of AmpH genes in M. leprae as compared to M. tuberculosis and S. aureus. All AmpH1, AmpH2 and AmpH3 genes were pseudogenes in M. leprae but such sequences could be detected by BLAST-N search. But we did not able to detect AmpH4 gene fragment in M. leprae suggesting complete deletion. More interestingly, blaC gene was also silent in M. leprae whereas MmpL5 RND protein was also missing in M. leprae. The PonA1, PonA2, PbpA, PbpB and MecA_N proteins were very similar to M. tuberculosis (90% similarities) and penicillin resistance was mediated by these enzymes in M. leprae [78-80]. Interestingly, similar to TB, Leprosy treatment was never suggested by any β-lactam drug! By virtue of analysis, however, we predicted that M. leprae would be more sensitive to penicillin drugs. The analysis of bacterial PBP1B or PonA2 protein multi-alignment and phylogenetic analysis found M. leprae also association with S. aureus while M. tuberculosis, M. bovis, M. asiaticum, and M. canittii were in one group whereas M. avium, M. colombiens and M. intracellulare in a different group. Thus, sequence similarities among the blaAmpH genes of different Mycobacterium species found well but quite different than E. coli related gram-negative bacteria or gram-positive bacteria like Vibrio, Streptococcus and Staphylococcus species. Surely, now overexpression-purification and characterization of AmpH proteins of M. tuberculosis to develop a new drug against TB is a priority area of research. Further, comparison of PBPs in other important pathogens was reported [81,82]. The phage therapy protocols were also progressing in mycobacterial diseases [83,84]. The phyto-drugs were another remedy protocol for TB and Mt RNA polymerase was inhibited by a terpentinepolybromophenol from Cassia fistula (golden showers) bark [85,86]. We discussed the PENEM drug resistance in Mt but multi-resistance against other TB specific drugs (rifampicin, dapsone, bedaquiline, isoniazid, moxifloxacin) were more complex [87-90].
Conclusion
So far ponA1, PonA2, PbpA, PbpB, DacB1 and DacB2 were implicated in penicillin-binding and hydrolysis in M. tuberculosis. Using bioinformatics BLAST homology approach, we first time showed that blaAmpH type four class-C β-lactamases similar to blaAmpC, blaDHA, blaACC and blaCMY could control the cephalosporin hydrolysis in Mt and such genes appeared as pseudogenes in M. leprae. The Mt AmpH enzymes had better homologies with V. parahaemolyticus or Yerdinia pekkanenii AmpH enzyme as well as E. coli AmpH enzyme. The blaC class-A β-lactamase has similarity to S. aureus blaZ β-lactamase as well as blaCARB gene of V. parahaemolyticus but such gene also inactivated in M. leprae including an important drug efflux gene called MmpL5. Thus, we showed a different new mechanism of PENEM drug resistance in M. tuberculosis and M. leprae. All primers for Mt PBPs were made to check the transcription of individual genes by RT-PCR as well as to check chromosomal location by BLASTN search after WGS to avoid the concurrent mistakes in the nomenclature of Mt PBPs. Interestingly, we showed that ISKpn6 transposase gene was involved in the genesis of blaAmpH4 gene suggesting the mechanism of chromosomal genesis of β-lactamases likely under high dose of penicillin and cephalosporin drugs.
Acknowledgment
We thank NCBI for free database and CLUSTAL-Omega free software. AKC is retired professor of Biochemistry. AKC thanks Prof. Bidyut Bandhopadhyay, Principal of OIST for his interest in Mycobacterium research. AKC also thanks Prof. Sujoy Dasgupta, retired chairman of microbiology department, Bost Institute, Kolkata for suggestion on tuberculosis research.
Conflict of Interest
The author declares no competent interest and the data is computer generated.
References
- Chakraborty AK. Current status and unusual mechanism of multi-resistance in Mycobacterium tuberculosis. J Health Med Informatics. 2019;10(1): 328.
- Chakaya J, Khan M, Ntoumi F, Aklillu E, Fatima R, Mwaba P, et al. Global tuberculosis report 2020-reflections on the global TB burden, treatment and prevention efforts. Int J Infect Dis. 2021;113:S7–S12.
[Crossref] [Google Scholar] [PubMed]
- World Health Organization. Molecular line probe assays for rapid screening of patients at risk of Multidrug-Resistant Tuberculosis (MDR-TB). Geneva: World Health Organization. 2008.
- World Health Organization/IUATLD. Global Project on anti-tuberculosis drug resistance surveillance, anti-tuberculosis drug resistance in the world. Publication no. WHO/TB/97.229. Geneva, Switzerland: World Health Organization. 1998.
- Palomino JC, Martin A. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiot. 2014;3:317–340.
[Crossref] [Google Scholar] [PubMed]
- Soltys MA. The effect of penicillin on Mycobacteria in vitro and in vivo. Tubercle. 1952;33(4):120–125.
- Kasik JE, Weber M, Winberg E, Barclay WR. The synergistic effect of dicloxacillin and penicillin G on murine tuberculosis. Am Rev Respir Dis. 1966;94(2):260–261.
[Crossref] [Google Scholar] [PubMed]
- Kumar G, Galanis C, Batchelder HR, Townsend CA, Lamichhane G. Penicillin binding proteins and b-lactamases of Mycobacterium tuberculosis: Re-examination of the historical paradigm. mSphere. 2022;7(1):e00039-22.
[Crossref] [Google Scholar] [PubMed]
- Wivagg CN, Bhattacharyya RP, Hung DT. Mechanisms of b-lactam killing and resistance in the context of Mycobacterium tuberculosis. J Antibiot. 2014;67:645–654.
[Crossref] [Google Scholar] [PubMed]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiology Reviews. 2008;32(2): 234–258.
[Crossref] [Google Scholar] [PubMed]
- Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, et al. Molecular detection of mutations associated with first and second-line drug resistance compared with conventional drug susceptibility testing of M. tuberculosis. Antimicrob Agents Chemother. 2011;55(5):2032–2041.
[Crossref] [Google Scholar] [PubMed]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104(6):901-912.
[Crossref] [Google Scholar] [PubMed]
- Garcia SR, Otal I, Martin C, Gomez-Lus R, Ainsa JA. Novel streptomycin resistance gene from Mycobacterium fortuitum. Antimicrob Agents Chemother. 2006;50:3920-3922.
[Crossref] [Google Scholar] [PubMed]
- Sreevatsan S, Pan X, Stockbauer KE. Characterization of RPSL and RRS mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob Agents Chemother. 1996;40(4):1024-1026.
[Crossref] [Google Scholar] [PubMed]
- Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, et al. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005; 308(5727):1480–1483. PMID:15933203.
[Crossref] [Google Scholar] [PubMed]
- WHO. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response, World Health Organization, Geneva, Switzerland.
- Islam MM, Tan Y, Hameed HMA, Liu Z, Chhotaray C. Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect. 2018;S1198-743X(18)30794-30798.
[Crossref] [Google Scholar] [PubMed]
- da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in M. tuberculosis: Classical and new drugs. J Antimicrob Chemother. 2011;66:1417–1430.
[Crossref] [Google Scholar] [PubMed]
- Madsen CT, Jakobsen L, Buriankova K, Doucet-Populaire F, Pernodet JL, Douthwaite S. Methyltransferase Erm(37) slips on rRNA to confer atypical resistance in Mycobacterium tuberculosis. J Biol Chem. 2005;280(47):38942–38947.
[Crossref] [Google Scholar] [PubMed]
- Zhang B. Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell. 2019;176:636–648.
[Crossref] [Google Scholar] [PubMed]
- Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3(6):E2375.
[Crossref] [Google Scholar] [PubMed]
- Alderwick LJ, Harrison J, Lloyd GS, Birch HL. The Mycobacterial cell wall-peptidoglycan and arabinoglycan. Cold Spring Harbor Perspect Med. 2015;5:a021113.
[Crossref] [Google Scholar] [PubMed]
- He XC, Zhang XX, Zhao JN. Epidemiological trends of drug-resistant tuberculosis in China from 2007 to 2014: A retrospective study. Medicine (Baltimore). 2016;95(15):e3336.
[Crossref] [Google Scholar] [PubMed]
- Duan Q, Chen Z, Chen C. The prevalence of drug-resistant tuberculosis in mainland China: An updated systematic review and meta-analysis. PLoS One. 2016;11(2): e0148041.
[Crossref] [Google Scholar] [PubMed]
- Vilcheze C, Jacobs WR Jr. The isoniazid paradigm of killing, resistance, and persistence in Mycobacterium tuberculosis. J Mol Biol. 2019;2836(19):S0022-30093.
[Crossref] [Google Scholar] [PubMed]
- Scorpio A, Zhang Y. Mutations in pncA gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nature medicine.1996;2(6):662–667.
[Crossref] [Google Scholar] [PubMed]
- Smith T, Wolff KA, Nguyen L. Molecular biology of drug resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol. 2013;374:53–80.
[Crossref] [Google Scholar] [PubMed]
- Uchiya K, Takahashi H, Nakagawa T, Yagi T, Moriyama M. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium subsp. hominissuis. PLoS One. 2015;10(2):e0117797.
[Crossref] [Google Scholar] [PubMed]
- Matsumoto CK, Bispo PJ, Santin K, Nogueira CL, Leao SC. Demonstration of plasmid-mediated drug resistance in Mycobacterium abscessus. J Clin Microbiol. 2014 ;52(5):1727-1729.
[Crossref] [Google Scholar] [PubMed]
- Chakraborty AK. Poor correlation of diversified MDR genes in Gonococci plasmids: Does alteration in chromosomal DEGs, PBP2 and target mutations sufficient to widespread multi-resistance in Neisseria gonorrhoeae? J Health Med Informat. 2018;9:310.
- Chakraborty AK, Maity M, Patra S, Mukherjee S, Mandal T. Complexity, heterogeneity and mutational analysis of antibiotic inactivating acetyl transferases in MDR conjugative plasmids conferring multi-resistance. Res Rev: J Microbiol Biotechnol. 2017;6(2):28-43.
- Das S, Datta S, Dey M, Poria K, Chakraborty AK. Green Chemistry: Phyto-antibiotics, A green antibiotics, isolated from medicinal plant of West Bengal targeting multi-drug resistant bacteria". Acta Scientific Medical Sciences, 2022;6(5):127-143.
- Chakraborty AK. Complexity, heterogeneity, 3-D structures and transcriptional activation of multi-drug resistant clinically relevant bacterial beta-lactamases. Trends Biotechnol-open access. 2016;2(1):1-001.
- Jacoby GA. AmpC β-lactamases. Clini Microbiol Rev. 2009;22(1):161-182.
- Chakraborty AK, Pradhan S, Das S, Maity M, Sahoo S, Poria K. Complexity of OXA Beta-Lactamases involved in multi-resistance. British J Bio-Medical Res. 2019;3(1):772-798.
- Chakraborty AK, Poira K, Saha D, Halder C, Das S. Multidrug-resistant bacteria with activated and diversified MDR genes in Kolkata Water: Ganga action plan and heterogeneous phyto-antibiotics tackling superbug spread in India. American J Drug Deli Therapeutics. 2018;5(1):2.
- Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae 1929. Bull World Health Organ. 2001;79:780–790.
[Google Scholar] [PubMed]
- Tan SY, Tatsumura Y. Alexander Fleming (1881-1955): Discoverer of penicillin. Singapore Med J. 2015;56(7):366-377.
[Crossref] [Google Scholar] [PubMed]
- Lynch JP, Clark NM, Zhanel GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum beta-lactamases and carbapenemases). Exp Opini Pharmacother. 2013;14:199-210.
[Crossref] [Google Scholar] [PubMed]
- Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37-67.
[Crossref] [Google Scholar] [PubMed]
- Goffin C, Ghuysen JM. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62(4):1079-1093.
[Crossref] [Google Scholar] [PubMed]
- Bhakta S, Basu J. Overexpression, purification and biochemical characterization of a class A high-molecular-mass Penicillin-Binding Protein (PBP), PBP1* and its soluble derivative from Mycobacterium tuberculosis. Biochem J. 2002;361:635-639.
[Crossref] [Google Scholar] [PubMed]
- Voladri RK, Lakey DL, Hennigan SH, Menzies BE, Edwards KM, Kernodle DS. Recombinant expression and characterization of the major beta-lactamase of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1375–1381.
[Crossref] [Google Scholar] [PubMed]
- Sung MT, Lai YT, Huang CY, Chou LY, Shih HW, Cheng WC, et al. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc Natl Acad Sci USA, 2009;106:8824-8829.
[Crossref] [Google Scholar] [PubMed]
- Schiffer G, Holtje JV. Cloning and characterization of PBP 1C, a third member of the multi-modular class A penicillin-binding proteins of Escherichia coli. J Biol Chem. 1999;274:32031-32039.
[Crossref] [Google Scholar] [PubMed]
- Diene SM, Pontarotti P, Azza S, Armstrong N, Pinault L, Chabriere E, et al. Origin, diversity, and multiple roles of enzymes with metallo-β-lactamase fold from different organisms. Cells. 2023;12(13):1752.
[Crossref] [Google Scholar] [PubMed]
- Gonzalez-Leiza SM, de Pedro MA, Ayala JA. AmpH, a bifunctional DD-endopeptidase and DD-carboxypeptidase of Escherichia coli. J Bacteriol. 2011;193(24):6887-6894.
[Crossref] [Google Scholar] [PubMed]
- Henderson TA, Young KD, Denome SA, Elf PK. AmpC and AmpH, proteins related to the class C beta-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J Bacteriol. 1997;179(19):6112-6121.
[Crossref] [Google Scholar] [PubMed]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res.1 988;16(22):10881-10890.
[Crossref] [Google Scholar] [PubMed]
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:539.
[Crossref] [Google Scholar] [PubMed]
- Altschul SF, Gish W, Miller W, Myers EM, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410.
[Crossref] [Google Scholar] [PubMed]
- Cole ST, Eiglmeier K, Parkhill J. Massive gene decay in the leprosy bacillus. Nature, 2001;409:1007-1011.
[Crossref] [Google Scholar] [PubMed]
- Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D. Infectious diseases society of America guidance on the treatment of Ampc Β-Lactamase-producing enterobacterales, carbapenem-resistant Acinetobacter baumannii, and Stenotrophomonas maltophilia Infections. Clin Infect Dis. 2022;74(12):2089-2114.
[Crossref] [Google Scholar] [PubMed]
- Jaurin B, Grunstrom T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901.
[Crossref] [Google Scholar] [PubMed]
- Lim D, Strynadka NC. Structural basis for the beta-lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol. 2002;9:870-876.
[Crossref] [Google Scholar] [PubMed]
- Pares S, Mouz N, Petillot Y, Hakenbeck R. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat Struct Biol. 1996;3:284-289.
[Crossref] [Google Scholar] [PubMed]
- Rice LB, Desbonnet C, Tait-Kamradt A, Garcia-Solache M, Lonks J. Structural and regulatory changes in PBP4 trigger decreased β-lactam susceptibility in Enterococcus faecalis. mBio. 2018;9(2):e00361-e003618.
[Crossref] [Google Scholar] [PubMed]
- Basuino L, Jousselin A, Alexander JAN, Strynadka NCJ, Pinho MG. PBP4 activity and its overexpression are necessary for PBP4-mediated high-level β-lactam resistance. J Antimicrob Chemother. 2018;73(5):1177-1180.
[Crossref] [Google Scholar] [PubMed]
- Poirel L, Guibert M, Girlich D. Cloning, sequence analyses, expression, and distribution of ampC ampR from Morganella morganii clinical isolates. Antimicrob Agents chemther. 1999;43:769-776.
[Crossref] [Google Scholar] [PubMed]
- Nadler JP, Berger J, Nord JA, Cofsky R, Saxena M. Amoxicillin-clavulanic acid for treating drug-resistant Mycobacterium tuberculosis. Chest. 1991;99:1025–1026.
[Crossref] [Google Scholar] [PubMed]
- Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–469.
[Crossref] [Google Scholar] [PubMed]
- Zhang J, Yang YH, Jiang YL, Zhou CZ, Chen Y. Structural and biochemical analyses of the Streptococcus pneumonia L,D-carboxypeptidase DacB. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 2):283-292.
[Crossref] [Google Scholar] [PubMed]
- Satishkumar N, Alexander JA, Poon R, Buggeln E, Argudín MA, Strynadka NC, et al. PBP4-mediated β-lactam resistance among clinical strains of Staphylococcus aureus. J Antimicrob Chemother. 2021;76(9):2268-2272.
[Crossref] [Google Scholar] [PubMed]
- da Costa TM, de Oliveira CR, Chambers HF, Chatterjee SS. PBP4: A new perspective on Staphylococcus aureus β-lactam resistance. Microorganisms. 2018;6(3):57.
[Crossref] [Google Scholar] [PubMed]
- Young M, Walsh DJ, Masters E. Identification of Staphylococcus aureus Penicillin Binding Protein 4 (PBP4) Inhibitors. Antibiotics (Basel). 2022;11(10):1351.
[Crossref] [Google Scholar] [PubMed]
- Matsuda K, Fujita K, Wakimoto T. PenA, a penicillin-binding protein-type thioesterase specialized for small peptide cyclization. J Ind Microbiol Biotechnol. 2021;48(3-4):kuab023.
[Crossref] [Google Scholar] [PubMed]
- Ohnishi M, Watanabe Y, Ono E, Takahashi C, Oya H. Spread of a chromosomal cefixime-resistant penA gene among different Neisseria gonorrhoeae lineages. Antimicrob Agents Chemother. 2010;54(3):1060-1067.
[Crossref] [Google Scholar] [PubMed]
- Wivagg CN, Wellington S, Gomez JE, Hung DT. Loss of a class A penicillin-binding protein alters β-lactam susceptibilities in Mycobacterium tuberculosis. ACS Infect Dis. 2016;2(2):104-110.
[Crossref] [Google Scholar] [PubMed]
- Deshpande D, Srivastava S, Chapagain M. Ceftazidime-avibactam has potent sterilizing activity against highly drug-resistant tuberculosis. Sci Adv. 2017;3(8):e1701102.
[Crossref] [Google Scholar] [PubMed]
- Hett EC, Chao MC, Rubin EJ. Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria. PLoS Pathog. 2010;6(7):e1001020.
[Crossref] [Google Scholar] [PubMed]
- Shigemura K, Shirakawa T, Massi N. Presence of a mutation in ponA1 of Neisseria gonorrhoeae in numerous clinical samples resistant to various beta-lactams and other, structurally unrelated, antimicrobials. J Infect Chemother. 2005;11(5):226-230.
[Crossref] [Google Scholar] [PubMed]
- Dekhil N, Mardassi H. Genomic changes underpinning the emergence of a successful Mycobacterium tuberculosis Latin American and Mediterranean clonal complex. Front Microbiol. 2023;14:1159994.
- Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 74:1686-1701.
[Crossref] [Google Scholar] [PubMed]
- Kocaoglu O, Carlson EE. Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob Agents Chemother. 2015;59(5):2785-2790.
[Crossref] [Google Scholar] [PubMed]
- Kwon HH, Tomioka H, Saito H. Distribution and characterization of beta-lactamases of mycobacteria and related organisms. Tuber Lung Dis. 1995;76(2):141-148.
[Crossref] [Google Scholar] [PubMed]
- Kasik JE, Peacham L. Properties of beta-lactamases produced by three species of mycobacteria. Biochemical J. 1968;107:675-682.
[Crossref] [Google Scholar] [PubMed]
- Sugawara-Mikami M, Tanigawa K, Kawashima A. Pathogenicity and virulence of Micobacterium leprae. Virulenc. 2022;13:1985-2011.
[Crossref] [Google Scholar] [PubMed]
- Jones CH, Ruzin A, Tuckman M. Protocol for rapid determination of TEM and SHV-type extended-spectrum β-lactamases in clinical isolates and identification of the novel β-lactamase genes blaSHV-48, blaSHV-105, and blaTEM-155. Antimicrob Agents Chemother. 2009;53:9977-9986.
- Halim MZ, Jaafar MM, Teh LK. Genome sequencing and annotation of multidrug resistant Mycobacterium tuberculosis (MDR-TB) PR10 strain. Genom Data. 2016;7:245-246.
[Crossref] [Google Scholar] [PubMed]
- Blattner FR, Plunkett III G, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453-1462.
[Crossref] [Google Scholar] [PubMed]
- Ropp PA, Hu M, Olesky M. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2002;46:769-777.
[Crossref] [Google Scholar] [PubMed]
- Leski TA, Tomasz A. Role of Penicillin-Binding Protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol. 2005;187:1815-1824.
[Crossref] [Google Scholar] [PubMed]
- Guerrero-Bustamante CA, Dedrick RM, Garlena RA, Russell DA, Hatfull GF. Toward a phage cocktail for tuberculosis: Susceptibility and tuberculocidal action of mycobacteriophages against diverse Mycobacterium tuberculosis strains. mBio. 2021; 12: e00973- e00921.
[Crossref] [Google Scholar] [PubMed]
- Froman S, Will DW, Bogen E. Bacteriophage active against virulent Mycobacterium tuberculosis. I. Isolation and activity. Am J Public Health Nations Health, 1954;44:1326–1333.
[Crossref] [Google Scholar] [PubMed]
- Jiang RH, Xu HB, Fu J. Outcomes of Chinese herb medicine for the treatment of multidrug-resistant tuberculosis: A systematic review and meta-analysis. Complement. Ther Med. 2015;23(4):544-554.
[Crossref] [Google Scholar] [PubMed]
- Chakraborty AK, Saha S, Poria K. A saponin-polybromophenol antibiotic (CU1) from Cassia fistula bark targeting RNA polymerase. Current Research Pharmacol and Drug Discovery, 2022;3:100090.
[Crossref] [Google Scholar] [PubMed]
- Kadura S, King N, Nakhoul M. Systematic review of mutations associated with resistance to the new and repurposed Mycobacterium tuberculosis drugs bedaquiline, clofazimine, linezolid, delamanid and pretomanid. J Antimicrob Chemother. 2020;75(8):2031-2043.
[Crossref] [Google Scholar] [PubMed]
- Xu Y, Jia H, Huang H, Sun Z, Zhang Z. Mutations found in embCAB, embR, and ubiA genes of ethambutol-sensitive and-resistant Mycobacterium tuberculosis clinical isolates from China. Biomed Res Int. 2015;2015:951706.
[Crossref] [Google Scholar] [PubMed]
- Zheng R, Li Z, He F, Liu H, Chen J. Genome-wide association study identifies two risk loci for tuberculosis in Han Chinese. Nat Commun. 2018;9:4072.
[Crossref] [Google Scholar] [PubMed]
- Matsumoto CK, Bispo PJ, Santin K, Nogueira CL, Leao SC. Demonstration of plasmid-mediated drug resistance in Mycobacterium abscessus. J Clin Microbiol. 2014;52(5):1727-1729.
[Crossref] [Google Scholar] [PubMed]
Citation: Chakraborty AK (2023) Four Chromosomally Clustered AmpH Class-C ?-Lactamases Control the Cephalosporin Hydrolysis in Mycobacterium tuberculosis and Similar Genetic Loci Appeared as Pseudogenes in Mycobacterium leprae. Adv Pharmacoepidemiol Drug Saf. 13:374.
Copyright: © 2023 Chakraborty AK. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.