Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2020) Volume 11, Issue 7
First Record of Melanospora chionea as a Possible Cause of Pink Root Rot Disease on Tomato Plants in Egypt
Farag MF*Received: 01-Jun-2020 Published: 16-Jul-2020, DOI: 10.35248/2157-7471.20.11.500
Abstract
Tomato is one of the most important vegetable crops in the world. It is infected with several disease through the growth season, but new disease appeared as a new challenge to tomato productivity, causing pink root rot. Symptoms of pink root rot were observed on tomato (Lycopersicon esculentum Mill.) grown in Beni Sweif Governorate (Nasser, Sumosta, Beba and El-Wasta Counties) in summer 2013 as poor growth, chlorosis and then necrosis of the tip branches, by maturity. Typical symptoms on the infected root especially, epidermis were picked areas and both of cortex and vascular bundles were colored with pink along the infected tissues consistent with both those that were observed in the field. Based on morphological characteristics of the isolated fungus, disease symptoms and a pathogenicity test, Melanospora chionea was identified as the causal agent of pink root rot of tomato. Identification of this species was confirmed by sequencing of internal transcribed space (ITS region) of ribosomal RNA gene. M. chionea has not previously been reported on tomato. The host range of this disease was defined between numerous hosts belonging to Fabaceae, Malvaceae, Cucurbitaceae and Solanaceae. The aim of this work to determine and description of the disease and identification of the pathogen morphologicaly and genetically. More work is required to find appropriate methods for controlling this new disease.
Keywords
Melanospora chionea; Pink root rot; Morphological characteristics; Pathogenicity test; Lycopersicon esculentum; Egypt
Introduction
The genus Melanospora is characterized by ostiolate ascomata having a thin translucent peridium and clavate prototunicate asci which deliquesce when mature [1]. Morphologically, species of Melanospora are similar to members of Chaetmiaceae and the Sordariaceae, but they have Diaporthe type centrum [2]. Most species of Melanospora are parasitic on or closely associated with other fungi. They have a wide host range including Basidiomycetes and both sexual and asexual Ascomycetes. The wilt disease fungus Fusarium oxysporum is one of the most common hosts, and some Melanospora species fuse with the host protoplasts to obtain nutrients, an interaction called fusion biotrophism [3].
Doguet [4] listed forty- five species of Melanospora and regarded others as synonyms. With the aid of the scanning electron microscope (SEM), [5] further restricted the range of this genus based on ascospore morphology. Further species were added to the complex by Garcı´a et al. [6,7], Hawksworth et al. [8], Krug [9], Nitzan et al. [10], Stchigel et al. [11,12] and Vujanovic and Goh [13]. Chaudhary et al. [14] published the new genus Vittatispora from India, which also belongs to the Melanospora complex, and the non-sporulating Papulaspora was demonstrated to represent sterile Melanospora ascomata by Davey et al. [15]. Li Fan et al. [16] published a new species of Melanospora parasitizing Chinese black and white truffles (Melanospora subterranean).
In this paper, the role of M. chionea as a potential pathogen causing pink root rot disease of tomato plants is demonstrated. Morphological identification of the fungus was confirmed by both microscopic images and performing gene sequencing of rDNA.
Materials and Methods
For isolation from tomato root plants which collected from Beni Serif governorate (latitude and longitude at 29.0667 N and 31.0833 E), the roots were washed under tap water, chopped into 2 cm small pieces and surface sterilized in 0.5% NaOCl for two minutes then rinsed three times with sterilized distilled water, dried between sterilized filter paper and placed on PDA medium and finally kept in an incubator at 25 ± 2°C under dark conditions.
For maintaining M. chionea with a highly and rapidly ascus fruits, the fungus was cultured by placing small agar blocks containing single-ascospore on V8 juice vegetable medium, incubated at 30°C for 14 days. The mycelia and spores were then scraped from the surface, suspended in water and sieved through 3 layers of cheesecloth under aseptic conditions. The number of colony forming units (CFU) in the spore suspension was counted using a haemocytometer (Thoma, Germany) and the concentration was adjusted using sterilized tap water for pathogenicity tests.
Molecular identification of fungal isolates
Fungal isolates were grown in sterile Petri plates containing autoclaved Czapek`s Dox agar provided with 0.5% yeast extract (CYA) medium and incubated for 7 days at 28°C [17]. Cultures were sent to the Molecular Biology Research Unit, Assiut University for DNA extraction using Patho-gene-spin DNA/RNA extraction kit provided by Intron Biotechnology Company, Korea. The fungal DNA was then sent to SolGent Company, Daejeon South Korea for polymerase chain reaction (PCR) and rRNA gene sequencing. PCR was performed using ITS1 (forward) and ITS4 (reverse) primers which were incorporated in the reaction mixture. Primers have the following composition: ITS1 (5' - TCC GTA GGT GAA CCT GCG G - 3'), and ITS4 (5'- TCC TCC GCT TAT TGA TAT GC -3'). The purified PCR products (amplicons) were sequenced with the same primers [18].
The ITS dataset included 15 sequences, three of which are Melanospora damnosa AUMC 14376 and M. chionea AUMC 14438 and AUMC 14440 and 11 are sequences for the nearest strains of Melanospora downloaded from GenBank and including the available type species, in addition to sequence for Sarocladium implicatum as an out group. of Melanospora damnosa AUMC 14376 and M. chionea AUMC 14438 and AUMC 14440 were(DNA star version 5.05). Assembled sequences of the three Melanospora strains obtained in this study were aligned with those downloaded from GenBank using MAFFT [19,20]. Maximum-likelihood (ML) and Maximum parsimony (MP) phylogenetic analyses were performed using PhyML 3.0 [21] The robustness of the most parsimonious trees was evaluated by 100 bootstrap replications [22]. The best optimal model of nucleotide substitution for the ML analyses was determined using alkaline information criterion (AIC) as implemented in Model test 3.7 [23]. The phylogenetic tree was visualized using BioNJ [24], and edited using Fig. tree version 1.4.3 [25].
Pathogenicity test
The pathogenic capabilities of the isolated fungi were determined using seeds of tomato commercial cultivar (super jacal). In summer 2018, seeds of tomato were sown in plastic pots (30 cm) having sterile moist sand and were placed in greenhouse and watered as needed. When the seedlings reached the 15-25 cm tall with 3-5 true leaves they were gently removed from pots and the roots were dipped individually in a spore suspension (106 spores/ml) of the five isolates obtained previously from different counties of Beni Sweif governorate for 30 min. the inoculated seedlings were then transplanted in a potting mix consisting of clay, sand and peat (1:1:1). Seedlings were dipped in tap water used as control. The pots were placed in greenhouse and arranged as a randomized complete block design with three replicates. Three months after inoculation, the plants were assessed for disease symptoms and gently uprooted and the roots examined for signs of infection.
Host range
Host range studies were conducted on fourteen crop plants from six botanical families (Molvaceae, Asteraceae, Brassicaceae, Cucurbitaceae, Fabaceae and Solanaceae) were selected for testing. Sterile soil in pots (20 cm) was inoculated with ascospores suspension of a single isolate of M. chionea immediately after incubated at 30°C for 10-15 days on V8 medium to allow colonization. Ascospores concentrations as determine by dilution at 106 /ml. seeds of the host range plants were soaked 1hr in an ascospores suspension and then planted in the pots (3 replicates) previously colonized with the same isolate. Plants were grown in a greenhouse for 8-10 weeks. Plants were then uprooted to observe the pink root rot symptoms.
Results and Discussion
Symptoms on inoculated plants appeared as poor growth, chlorosis and then necrosis of the tip branches, by maturity. Typical symptoms on the infected root especially, epidermis were picked areas and both of cortex and vascular bundles were colored with pink along the infected tissues consistent with both those that were observed in the field, and those reported for this disease (Figures 1 and 2). Koch’s Postulates were fulfilled by reisloation of the tested fungus.
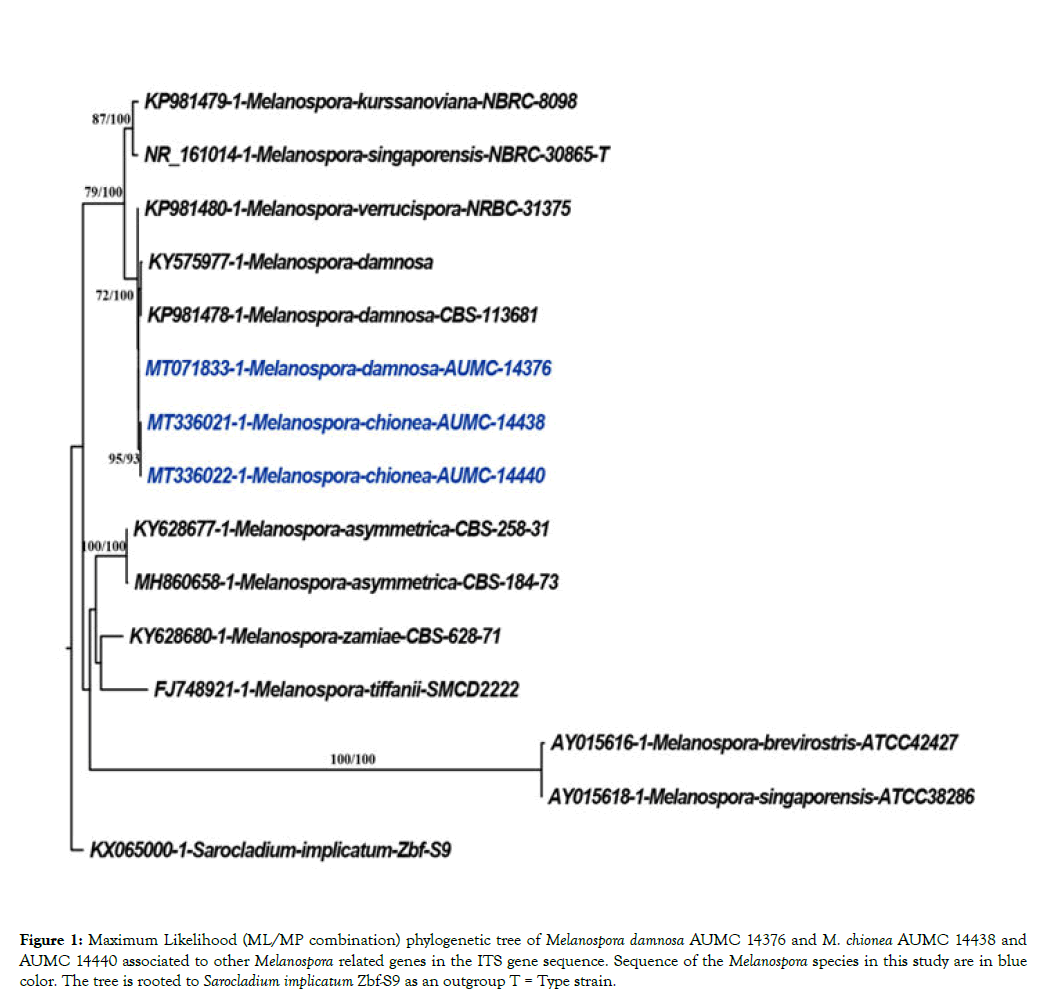
Figure 1: Maximum Likelihood (ML/MP combination) phylogenetic tree of Melanospora damnosa AUMC 14376 and M. chionea AUMC 14438 and AUMC 14440 associated to other Melanospora related genes in the ITS gene sequence. Sequence of the Melanospora species in this study are in blue color. The tree is rooted to Sarocladium implicatum Zbf-S9 as an outgroup T = Type strain.
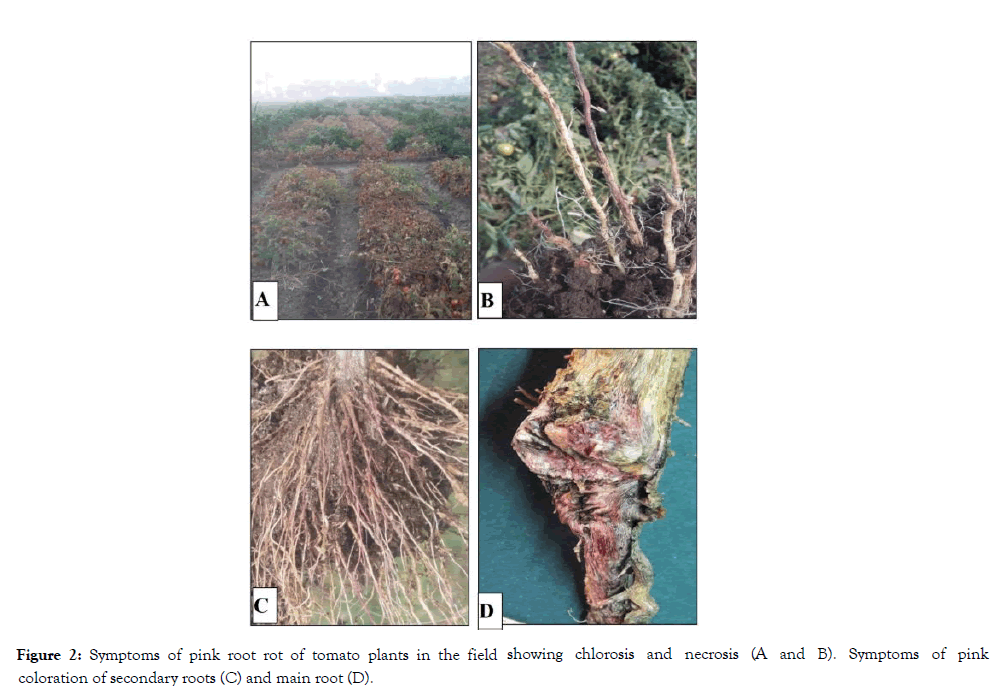
Figure 2: Symptoms of pink root rot of tomato plants in the field showing chlorosis and necrosis (A and B). Symptoms of pink coloration of secondary roots (C) and main root (D).
The fungus involved in the pink root rot of tomato was identified morphologically on the basis of perthecial structure, dimensions, shape and measurements of ascospores. The mycokeys provided by Cannon and Hawksworth [5] and Marin- Flex [19] were consulted for identification of the isolated fungi with the help of mycologists at Assiut University Mycological Center (AUMC). Observations made with bright field light microscopy showed that, perithecia are superficial, solitary to gregarious, sub-globose to globose yellow to pale brown. Beakʼs length 120-130 μm, slightly tapering, with a terminal ring of hyaline setae.
Ascospores are ellipsoidal to discoid, walls dark brown, with two terminal sessile pores measuring 14-15 × 10-11 μm as shown in Figures 3, 4 and 5. Images taken by scanning electron microscope (SEM) are illustrated in Figure 4. Microscopic observations of the fungus were consistent with Melanospora chionea described by Cannon and Hawksworth [5] and Marin- Flex et al. [19] To the best of our knowledge, this is the first published report of pink root rot of tomato caused by M. chionea. Sequencing data of ITS region of rDNA extracted from the two fungal strains (Table 1 and Figure 1) revealed that they are closely related to Melanospora damnosa showing 98.20% similarity with M. damnosa strain KY575977 isolated from triennial branches stems of stumped hazelnuts. It also showed 98.89% identity with M. damnosa AUMC14376 (MT071833) recovered from Phaseolus vulgaris leaves and 98.19% similarity with M. damnosa CBS 113681 (KP981478).

Figure 3: Mature perithecia 100x (A), Perithecia with ascospores 200x (B), perithecial beak with setae and discharging ascospores 400x (C). Higher magnification of ascospores 1000x (D).
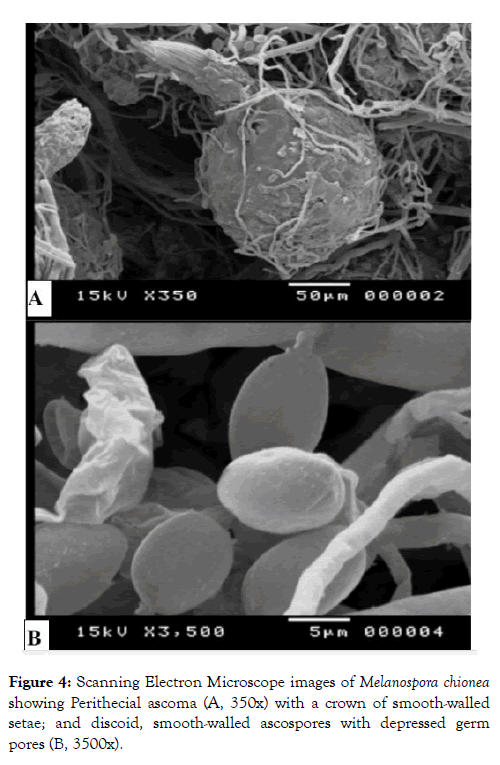
Figure 4: Scanning Electron Microscope images of Melanospora chionea showing Perithecial ascoma (A, 350x) with a crown of smooth-walled setae; and discoid, smooth-walled ascospores with depressed germ pores (B, 3500x).
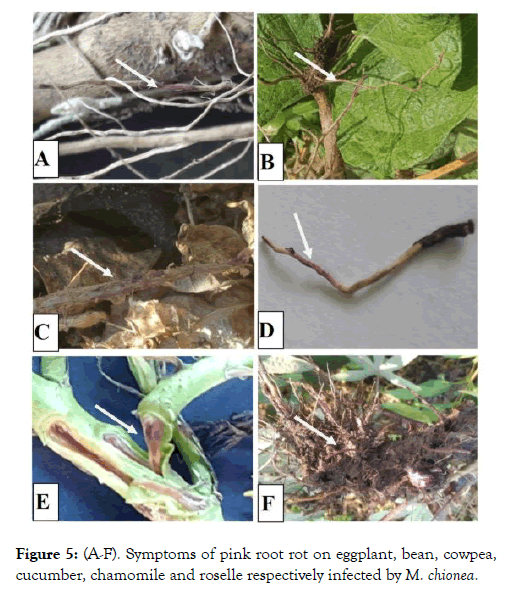
Figure 5: (A-F). Symptoms of pink root rot on eggplant, bean, cowpea, cucumber, chamomile and roselle respectively infected by M. chionea.
| Fungal species isolated in the present study | Closest strains accessed from the GenBank | |||||
|---|---|---|---|---|---|---|
| Name | Strain No. | Accession No. | Identity (%) | Coverage (%) | Source | |
| Melanospora chionea AUMC14438 (gb: MT336021) Melanospora chionea AUMC14440 (gb: MT336022) |
Melanospora damnosa | ------- | KY575977 | 98.41 | 99 | Stem of stumped hazelnut, China |
| Melanospora damnosa | CBS113681 | KP981478 | 98.05 | 99 | Spain | |
| Melanospora damnosa | AUMC14376 | MT071833 | 99.04 | 91 | Blighted seedlings of Phaseolus vulgaris, Egypt | |
| Melanospora verrucispora | NRBC31375 | KP981501 | 98.41 | 99 | Spain | |
Table 1: Percentage identity and coverage of ITS sequences of Melanospora species accessed from GenBank showing close relationship with M. chinea obtained in the current study.
The entire ITS data set comprised 15 sequences. The maximum parsimony dataset consisted of 583 characters with 405 constant characters (no gaps, no N), 324 variable characters which were parsimony-uninformative (80.0% of constant characters), and 295 characters were counted as parsimony informative (72.8% of constant). The GTR was the best fit model for nucleotides substitution. Maximum Parsimony analyses resulted in 2 most parsimonious trees with a tree length of 593 steps. Maximum likelihood analysis yielded one tree (Log-likelihood= -3852.02044) with a tree size of 4.77719.
However, the morphological characteristics of M. chionea showed some variations from M. damnosa particularly in the shape and dimensions of ascospores. In case of M. chionea the ascospores are smooth walled, discoid to ellipsoidal measuring 7.5-16 × 6-12 × 4-7 μm but those of M. damnosa are citriform to rhomboidal with larger dimensions measuring 18-28 × 10-14 μm [5]. Melanospora verrucispora showed also closer relationship (98.06% identity) with M. chionea but the distinct verrucose ascospores of M. verrucispora separates it from M. chionea [26].
Sequences of Melanospora chionea AUMC14438 (MT336021) (length 566bp)
GTAGGTGAAACTGCGGAGGGATCATTACAGAATCTGA CCATTTGTGTCATGGCTCTGCCAACCCTGTGAACTTTA TATACTTGTACGTTGCCTCGGCGGAACCTGCCTTTTTG GCAGGCCGCCGG
CCGGCATATACGCAAACGCTCTGAAAAAGCTCCGCGC TCTATCTGAATAAGAAAACTTTAACGAGTAAAAACTTTT GGCAACGGATCTCTTGGCTCTGGCATCGATGAAGAAC GCAGCGAAATG
CGATACGTAGTGTGAATTGCAGAATTCAGTGAACCATC GAATCTTTGAACGCACCTTGCGGCCGCCGGTAATCCG GCGGCCATGCCCGTCCGAGCGTCGTTTCGACCCTCG GGAGTTCTCCTCG
CAAGAGAAGTTCTCCCGGCCTTGGGCCAGCGTGTTGC GCGGCCCGCAAACCAGCGGCGGCAGTACCGGCGATG TCCTCTGTGCAGTAGATTTATAGAAAACTCGCATTGGT CTCCGGTAAGGC
TTGCCTTACAACCAACTTCTTTAGGTCGACCTCGGATC GGGTAGGGATACCCGCTAAACTTAAGCATATC
Melanospora chionea AUMC14440 (gb: MT336022) (length 568 bp)
CTGCGGAGGGATCATTACAGAATCTGACCATTTGTGTC ATGGCTCTGCCAACCCTGTGAACTTATATACTTGTACG TTGCCTCGGCGGAACCTGCCTTTTTGGCAGGCCGCC GGCCGGCATATAC
GCAAACGCTCTGAAAAAGCTCCGCGCTCTATCTGAAT AAGAAAACTTTAACGAGTAAAAACTTTTGGCAACGGAT CTCTTGGCTCTGGCATCGATGAAGAACGCAGCGAAAT GCGATACGTAG
TGTGAATTGCAGAATTCAGTGAACCATCGAATCTTTGA ACGCACCTTGCGGCCGCCGGTAATCCGGCGGCCATG CCCGTCCGAGCGTCGTTTCGACCCTCGGGAGTTCTCC TCGCAAGAGAAG
TTCTCCCGGCCTTGGGCCAGCGTGTTGCGCGGCCCG CAAACCAGCGGCGGCAGTACCGGCGATGTCCTCTGT GCAGTAGATTTATAGAAAACTCGCATTGGTCTCCGGTA AGGCTTGCCTTACA
ACCAACTTCTTTAGGTCGACCTCGGATCGGGTAGGGA TACCCGCTAAACTTAAGCATATCAATAAGCGGAGGA
Host range
Observations of the host range trials in Table 2 showed that tomato isolate was pathogenic on eggplant, common bean, pea, cowpea, watermelon, muskmelon, roselle, cabbage, chamomile and Snake cucumber beside tomato. Symptoms on all species included poor growth, chlorosis and then necrosis of the tip branches, by maturity. Typical symptoms on the infected root especially, epidermis were picked areas and both of cortex and vascular bundles were coloured with pink along the infected tissues (Figure 4).
| N | Family | Plant species, Common name |
Disease reaction |
|---|---|---|---|
| 1 | Solanaceae | Solanum melongena Linn Egg plant |
+ |
| Lycopersicon esculentumMill. Tomato |
+ | ||
| Capsicum annuum Linn. Pepper |
- | ||
| Solanum tuberosum Linn. Potato |
- | ||
| 2 | Fabaceae | Phaseolus vulgaris Common bean |
+ |
| Pisum sativum Pea |
+ | ||
| Vigna unguiculatasubsp. unguculata Cowpea |
+ | ||
| 3 | Cucurbitaceae | Cucumis melo Naud Muskmelon |
+ |
| Cucumis melo var. flexuosus Snake cucumber |
+ | ||
| Citrullus lanatus Watermelon |
+ | ||
| Cucumis sativusL. Cucumber |
- | ||
| 4 | Brassicaceae | Brassica oleracea var. Capitata Cabbage |
+ |
| 5 | Asteraceae | Matricaria chamomilla chamomile |
+ |
| 6 | Molvaceae | Hibiscus sabdariffa Roselle |
+ |
Table 2: Susceptibility of various species infection by Melanospora chionea.
However, the response of squash, potato and pepper showed no infection. Although, most species of Melanospora are parasitic on or closely associated with other fungi, but Melanospora subterranean is the first record of Melanospora species parasitizing Chinese black and white truffles (T. indicum and T. huidongense), and its host range indicates that the new species might be a disease threat to commercially exploited European truffles including Tuber melanosporum and Tuber magnatum. [16]. Also, M. zamiae, the causal agent of crown rust of maize was detected in only one accession (IC-587520) of C. speciosus from Giridih, Jharkhand. The report of the Technical Working Group 1 of Australia listed this as a pathogen of quarantine significance and highlighted the risk associated with the bulk maize import from USA [27].
Conclusion
Our results were similar with the previously on Chinese black and white truffles and maize as the pathogen but it differs in infection and symptoms. As the author is far aware, there is no available information on tomato pink root rot disease with Melanospora chionea in Egypt. Therefore it can be concluded that the present work is a first record of tomato pink root rot disease by M. chionea, so we will complete survey this disease in Egypt, epidemiology of this disease and we will harry up more required work to find appropriate methods for controlling this new disease.
Acknowledgement
I am especially grateful to Dr. M.F. Attia, Faculty of Agriculture, Cairo University for his kind help, moral support and wise guidance comments. Also I am greatly thankful to Dr. A.M. Moharram, Director of Assiut University Mycological Centre for checking morphological and molecular identification of the fungal strains.
REFERENCES
- Hanlin RT, Goh TK, Tortotera O. Icones Aascomycetum Venezuelae Melanospora zamiae. Mycotaxon. 1993;49:167-174.
- Luttrell ES. Taxonomy of the Pyrenomycetes. Univ. Missouri Stud. 1951;24:1-120.
- Jeffries P, Young TWK. Interfungal parasitic relationships. CAB International, Wallingford. 1994;60.
- Doguet G. Le genre Melanospora biologie, morphologie, development, systematique. Botaniste. 1955;39:1-313.
- Cannon PF, Hawksworth DL. A re-evaluation of Melanospora corda and similar pyrenomycetes, with a revision of the British species. Botanical Journal of the Linnean Society. 1982;84:115-160.
- Garcıa D, Stchigel AM, Guarro J. A new species of Syspastospora from tropical soils. Mycologia. 2002;94:862-865.
- Garcıa D, Stchigel AM, Guarro J. Two new species of Sphaerodes from Spanish soils. Stud Mycol 2004;50:63-68.
- Hawksworth DL, Henrici A, Kirk PM. Melanospora pegleri, a new species from fallen Laurus and other leaves in the British Isles. Kew Bull. 1999;54:795-799.
- Krug JC. A new species of Persiciospora from African soil. Mycologia. 1988;80:414-417.
- Nitzan N, Rogers JD, Johnson DA. Melanospora washingtonensis, a new species from potato. Sydowia 2004; 56:281-287.
- Stchigel AM, Guarro J, Figueras MJ. A new species of Melanospora from India. Mycologia 1996; 101:446-488.
- Stchigel AM, Cano J, Guarro J. A new species of Melanospora from Easter Island. Mycol Res. 1999;103:1305– 1308.
- Vujanovic V, Goh YK. Sphaerodes mycoparasitica sp. nov., a new biotrophic mycoparasite on Fusarium avenaceum, F. graminearum and F. oxysporum. Mycol Res. 2009;113:1172-1180.
- Chaudhary P, Campbell J, Hawksworth DL, Sastry KN. Vittatispora, a new melanosporaceous genus from Indian soil. Mycologia. 2006; 98:460-467.
- Davey ML, Tsuneda A, Currah RS. Evidence that the gemmae of Papulaspora sepedonioides are neotenous perithecia in the Melanosporales. Mycologia. 2008;100:626- 635.
- Li-Fan CH, Paul FC, Yu L. A new species of Melanospora on truffles from China. Mycologia. 2017;104:1433-1442.
- Kidd S, Halliday C, Alexiou H, Ellis D. Description of Medical Fungi Third edition National Mycology Reference Centre,Sa Pathology, Adelaide, Australia. 2016;1-266.
- White TJ, Bruns, T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A guide to Methods and Applications (ed. M. A. Innis D, Gelfand H, Sninsky JJ, White TJ. Academic Press: San Diego, USA. 1990;315-322..
- Katoh K, Standley DM, MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30:772-780.
- Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC evolutionary biology. 2010;10:210.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307-321.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783-791.
- Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817-818.
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular biology and evolution. 1997;14:685-695.
- Rambaut A, Drummond A. FigTree version 1.4. 0.2012.
- Marin-Felix Y, Guarro J, Cano-Lira JF, García D , Miller AN, Stchige AM. Melanospora (Sordariomycetes, Ascomycota) and its relatives. MycoKeys. 2018; 44: 81-122.
- Irwin J, Singh S, Kochman J, Murray G. Pathogen risks associated with bulk maize imports to australia from the United States of America. A report by technical working group 1: Disease risk analysis, for the import of maize from the USA for processing and use as animal feed. 1999;126.
Citation: Farag MF (2020) First Record of Melanospora chionea as a Possible Cause of Pink Root Rot Disease on Tomato Plants in Egypt. J Plant Pathol Microbiol. 11: 500. doi: 10.35248/2157-7471.20.11.500.
Copyright: © 2020 Farag MF. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

