Indexed In
- Open J Gate
- Genamics JournalSeek
- ResearchBible
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2019) Volume 8, Issue 1
Establishing a Remedy for Phenylketonuria Disease from Indian Ayurvedic Herbs
Preenon Bagchi1*, SophieVisvikis-Sieste1 and Ajit Kar22Satsang Herbal Research Laboratory, Satsang, Deoghar, Jharkhand, India
Received: 27-Mar-2019 Published: 31-May-2019, DOI: 10.35248/2167-7956.19.8.177
Abstract
The phenylketonuria (PKU) disease is an inherited disorder that increases the levels of a substance called phenylalanine in the blood and if not treated, phenylalanine can build up to harmful levels in the body. People with this disorder can't break down the amino acid phenylalanine. This phenylalanine, then builds up in the blood and brain causing intellectual disability and other serious health problems. It is rare but a serious inherited disorder. The main objective of this study is to establish a remedy for the phenylketonuria disease (novel drug leads for phenylketonuria disease’s receptors viz. ASCL1 gene (achaete-scute family bHLH transcription factor 1), GCH1 gene (GTP cyclohydrolase 1) and MAOB (Monoamine Oxidase B)) using phytocompounds from ayurvedic herbs. To achieve this objective we performed virtual screening with phytocompounds from ayurvedic herbs against the phenylketonuria disease’s receptors followed by ADME studies on the phytocompounds selected by virtual screening. Based on the analysis of the results of virtual screening and subsequent ADME studies on the phytocompounds it is seen that curcumin can be successfully considered as novel drug lead for treating phenylketonuria disease.
Keywords
Phenylketonuria; ADME; ASCL 1 gene; GHC1 gene; MAOB gene; Modeling; Docking
Introduction
Phenylketonuria which is commonly known as PKU, is an inherited disorder that increases the levels of phenylalanine (which is a building block of proteins) in the blood that is obtained through the diet (it is found in all proteins and in some artificial sweeteners) [1-3]. If PKU is not timely treated, phenylalanine can build up to harmful levels of toxins in the body, causing brain damage. The U.S. Food and Drug Administration (FDA) has approved the drug sapropterin dihydrochloride (Kuvan®) for the treatment of PKU. Kuvan® is a form of BH4, which is a substance in the body that helps break down phenylalanine [4-6]. PKU is caused by mutations in the gene PAH, GCH1, MAOB, ALB, IGF1, ASCL1, among others Koch et al., Moyle et al., Naz and John. PKU`s symptoms include seizures, tremors or trembling and shaking, stunted growth, hyperactivity, skin conditions such as eczema, a musty odor (bad smell) in their breath, skin or urine. Infants with classic PKU appear normal until they are a few months old and without treatment, these children develop permanent intellectual disability. Children with PKU usually have lighter skin and hair than unaffected family members and more prone to have skin disorders like eczema [1-9].
Genes considered in this work
Classical PKU is an autosomal recessive disorder caused by mutations in both alleles of the gene coding for phenylalanine hydroxylase found on chromosome 12.
ASCL1 gene (achaete-scute family bHLH transcription factor 1)
It encodes a member of the basic helix- loop-helix (BHLH) family of transcription factors, a protein that activates transcription by binding to the E box (5'- CANNTG-3'). Dimerization with other BHLH proteins is required for efficient DNA binding and this protein plays a role in the neuronal commitment and differentiation and in the generation of olfactory and autonomic neurons. Mutations in this gene cause the congenital central hypoventilation syndrome (CCHS) phenotype in rare cases [10].
GCH1 gene (GTP cyclohydrolase 1)
It provides instructions for making an enzyme called GTP cyclohydrolase 1, which is involved in the first of three steps in the production of a molecule called tetrahydrobiopterin (BH4). Other enzymes help to carry out the second and third steps in this process. Tetrahydrobiopterin plays a significant role in processing several protein building blocks (amino acids) in the blood; specifically, tetrahydrobiopterin, which is involved in the production of two neurotransmitters called dopamine and serotonin. Among their many functions, dopamine spreads signals within the brain to produce smooth physical movements and serotonin regulates mood, emotion, sleep, and appetite. Since it helps enzymes carry out chemical reactions, tetrahydrobiopterin is known as a cofactor [11].
MAOB (Monoamine oxidase B)
The protein coded by this gene belongs to the flavin monoamine oxidase family and it is an enzyme located in the mitochondrial outer membrane. It catalyzes the oxidative deamination of biogenic and xenobiotic amines and plays an important role in the metabolism of neuroactive and vasoactive amines in the central nervous system and peripheral tissues. This protein preferentially degrades benzylamine and phenylethylamine [12].
Ayurvedic herbs used and their active components
• Wood betony: The principal chemical components present in this plant are Tannins, Betulinic acid, oleonilic acid, rosamarinic acid, rutin, urosolic acid, stachydrine and glycosides.
• Nettle: The principal chemical components present in this plant are histamine, formic acid, acetylcholine, serotonin and vitamins.
• Plantago ovate: The principal chemical components present in this plant are xylose, arabinose, alanine, valine, glutamic acid, glycine, cysteine, lysine, leucine, tyrosine and xylose.
• Turmeric: The principal chemical components present in this plant are curcumin and camphene.
• Dandelion: The principal chemical components present in this plant are taraxacin, laevulin, resin and inulin.
The main objective of this study is to establish a novel ligand as drug for the PKU from the phytocompounds of the above mentioned ayurvedic herbs.
Methodology
The proteins corresponding to the genes for the PKU were downloaded from Genbank database and their 3d structures were modeled using modeller [1,13]. Modeller is used for homology or comparative modeling of protein three-dimensional structures wherein the user provides an alignment of a sequence to be modeled with known related structures and modeller automatically calculates a model containing all non-hydrogen atoms [13]. The modeller generated models were verified using Ramachandran Plot [1,14]. Ramachandran Plot is a way to visualize energetically allowed regions for backbone dihedral angles ψ against φ of amino acid residues in protein structure [14]. The 3d structures of the phytocompounds mentioned above were downloaded from pubchem database. These compounds were docked with the PKU receptors using PATCHDOCK server which is a server for molecular docking [1,15]. ADME studies were done with the phytocompounds which showed best docking results with the PKU receptors [1,16,17]. ADME is “absorption, distribution, metabolism, and excretion” which is the disposition of a pharmaceutical compound within an organism. This is based on Lipinski's rule of five to evaluate drug-likeness or determine if a chemical compound with a certain pharmacological or biological activity has chemical properties and physical properties that would make it a likely orally active drug in humans [17].
Results and Discussion
Homology modelling
PKU (phenylketonuria) gene receptors were retrieved from GENBANK database Table 1. The homologous templates of the receptors in Table 1 were selected using BLAST search against Protein Data Bank (PDB) and the selected templates were downloaded from PDB Table 2A-C. Using Modeller, the 3d structures of the receptors in Table 1 were modelled [13]. The models were verified using Rampage Ramachandran Plot server Table 3A-C and Figure 1A-C [14].
| Serial No. | Receptor Name | Code | Accession Number |
|---|---|---|---|
| 1 | Achaete-scute family bHLH transcription | ASCL1 | P50553.2 |
| 2 | GTP cyclohydrolase | GCH1 | P30793.1 |
| 3 | Monoamine oxidase B | MAOB | P27338.3 |
Table 1: PKU genes with Genbank accession number.
| Accession | Query cover | Identity |
|---|---|---|
| 2QL2 B | 22% | 45% |
| 4AYA A | 22% | 41% |
| 2YPAB* | 64% | 33% |
Homologous template search by https://toolkit.tuebingen.mpg.de/#/tools/hhpred since BLAST generated two homologous templates [18]
Table 2(A): Homologous template of ASCL1.
| Accession | Query cover | Identity |
|---|---|---|
| 1IS7 A | 92% | 93% |
| 1FB1 A | 78% | 100% |
| 1WM9 A | 74% | 58% |
Table 2(B): Homologous template of GCH 1.
| Accession | Query cover | Identity |
|---|---|---|
| 1GOS A | 100% | 100% |
| 2C73 A | 100% | 99% |
| 2BK4 A | 100% | 99% |
Table 2(C): Homologous template of MAOB.
| ASCL1 | ||||
|---|---|---|---|---|
| Number of residues in favored region (~98.0% expected) | Number of residues in allowed region (~2.0% expected) | Number of residues in outlier region | ||
| Model 1 | 184 (96.3%) | 5 (2.6%) | 2 (1.0%) | |
| Model 2 | 178 (93.2%) | 7 (3.7%) | 6 (3.1%) | |
| Model 3 | 177 (92.7%) | 12 (6.3%) | 2 (1.0%) | |
| Model 4 | 181 (94.8%) | 8 (4.2%) | 2 (1.0%) | |
| Model 5 | 185 (96.9%) | 5 (2.6%) | 1 (0.5%) | Selected |
Model 5 is selected as the best model since it has highest number of residues in the favored region and least number of residues in the outlier region.
Table 3(A-C): Ramachandran Plot Analysis of modeler generated models.
| GCH1 | ||||
|---|---|---|---|---|
| Number of residues in favored region (~98.0% expected) | Number of residues in allowed region (~2.0% expected) | Number of residues in outlier region | ||
| Model 1 | 235 (94.8%) | 7 (2.8%) | 6 (2.4%) | |
| Model 2 | 241 (97.2%) | 5 (2.0%) | 2 (0.8%) | Selected |
| Model 3 | 239 (96.4%) | 7 (2.8%) | 2 (0.8%) | |
| Model 4 | 237 (95.6%) | 8 (3.2%) | 3 (1.2%) | |
| Model 5 | 239 (96.4%) | 9 (3.6%) | 0 (0.0%) | |
Model 2 is selected as the best model since it has highest number of residues in the favored region
Table 3(B)
| MAOB | |||
|---|---|---|---|
| Number of residues in favored region (~98.0% expected) | Number of residues in allowed region (~2.0% expected) | Number of residues in outlier region | |
| Model 1 | 507(97.9%) | 9(1.7%) | 2(0.4%) Selected |
| Model 2 | 506(97.7%) | 10(1.9%) | 2(0.4%) |
| Model 3 | 506(97.7%) | 10(1.9%) | 2(0.4%) |
| Model 4 | 505(97.5%) | 11(2.1%) | 2(0.4%) |
| Model 5 | 506(97.7%) | 10(1.9%) | 2(0.4%) |
Model 1 is selected as the best model since it has highest number of residues in the favored region
Table 3 (C)
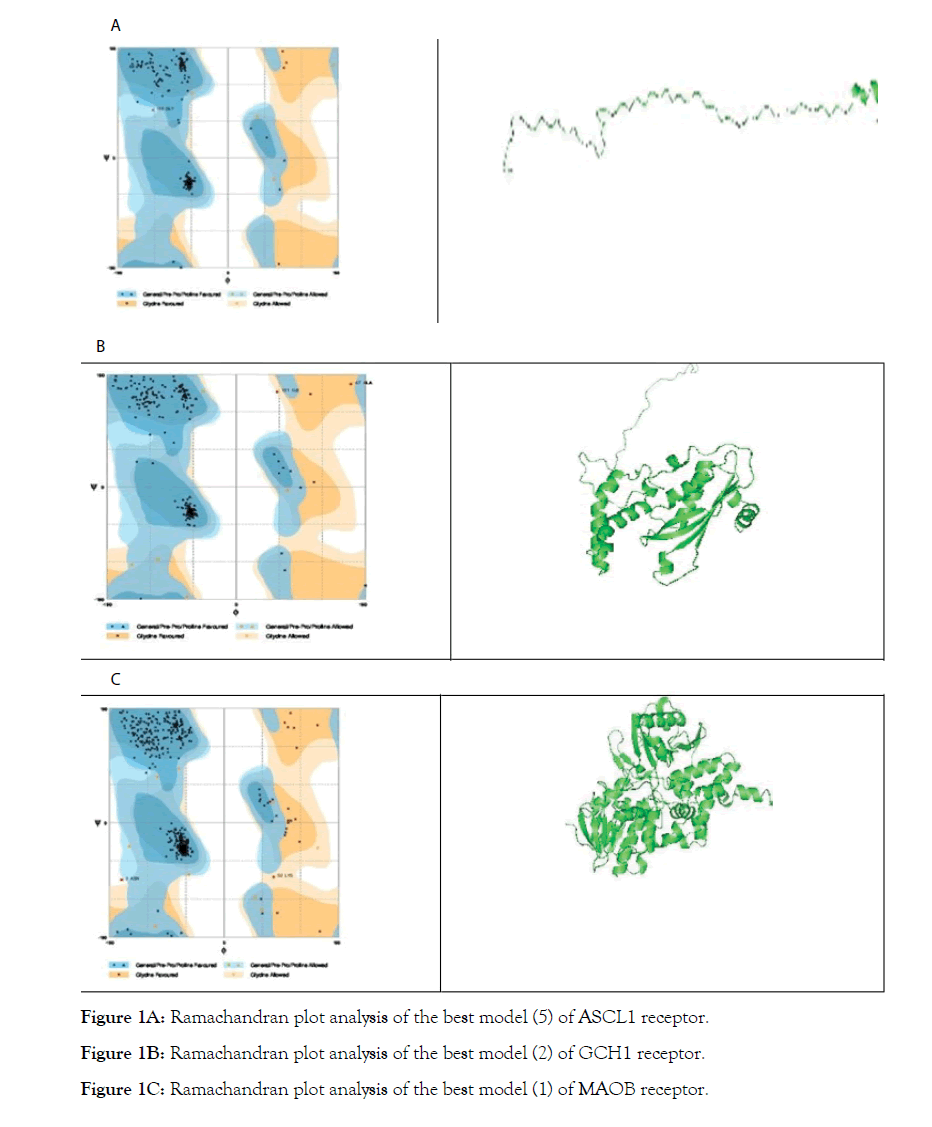
Figure 1A: Ramachandran plot analysis of the best model (5) of ASCL1 receptor.
Figure 1B: Ramachandran plot analysis of the best model (2) of GCH1 receptor.
Figure 1C: Ramachandran plot analysis of the best model (1) of MAOB receptor.
Docking
The selected models in Table 3 were docked with the phytocompounds from the Ayurvedic herbs using PATCHDOCK [15]. The docking scores were noted in Table 4A1-A5, B1- B5 and C1-C5. As per the virtual screening studies we find the phytocompounds betulinic acid, rutin, lecithin and curcumin docks with all the receptor genes. Hence we take these phycompounds into consideration (in comparison with the others used in this work) for further ADME studies to see which of these compounds show NO violations in the Lipinski rule of 5 (Figure 2A-C).
| WOOD BETONY | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | ASCL1 | Betulinic acid | 5640 | 5 | ASP-110 |
| 2 | ASCL1 | Delphinic acid | 2190 | 1 | SER-177 SER-144 GLU-182 TYR-193 |
| 3 | ASCL1 | Oleonilic acid | 5524 | 1 | PRO-143 |
| 4 | ASCL1 | Rosamarinic acid | 4642 | 2 | SER-148 |
| 5 | ASCL1 | Rutin | 5540 | 6 | GLU-180 |
| VAL-112 ALA-111 ASP-110 SER-156 |
|||||
Table 4 (A1): Docking results of ASCL1 receptor with compounds from Wood Betony.
| NETTLE | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | ASCL1 | Histamine | 2130 | 1 | TYR-193 |
| 2 | ASCL1 | Formic acid | 1190 | 2 | ARG-100 GLY-132 |
| 3 | ASCL1 | Acetylcholine | 2778 | 1 | ARG-131 |
| 4 | ASCL1 | Serotonin | 3202 | 3 | LYS-85 GLN-71 ARG-74 |
Table 4 (A2): Docking results of ASCL1 receptor with compounds from NETTLE.
| PLANTAGO OVATO | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | ASCL1 | Arabinose | 2182 | 3 | ARG-74 |
| 2 | ASCL1 | Xylose | 2360 | 4 | LEU-86 LYS-88 LEU-86 |
| 3 | ASCL1 | Valine | 2192 | 1 | ARG-74 LYS-84 LYS-88 ASN-67 |
| 4 | ASCL1 | Alanine | 1876 | 2 | GLY-132 |
| 5 | ASCL1 | Glutamic acid | 2280 | 4 | ARG-100 LYS-88 |
| 6 | ASCL1 | Glycine | 1582 | 3 | GLN-71 ASN-67 ARG-74 ARG-100 |
| 7 | ASCL1 | Cysteine | 1996 | 1 | GLY-132 GLU-90 ARG-131 |
| 8 | ASCL1 | Lysine | 2704 | 2 | ALA-179 ASP-110 |
| 9 | ASCL1 | Leucine | 2370 | 2 | ARG-74 ASN-67 |
| 10 | ASCL1 | Tyrosine | 2970 | 2 | SER-148 ASP-110 |
| 11 | ASCL1 | Rhamnose | 2418 | 3 | ASN-67 LYS-88 ARG-74 |
Table 4 (A3): Docking results of ASCL1 receptor with compounds from PLANTAGO OVATO.
| TURMERIC | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | ASCL1 | Curcumin | 5016 | 5 | VAL-125 ARG-126 SER-128 ALA-129 PRO-127 |
| 2 | ASCL1 | Camphene | 2862 | 0 | |
Table 4 (A4): Docking results of ASCL1 receptor with compounds from TURMERIC.
| DANDELION | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | ASCL1 | Taraxacin | 3922 | 2 | SER-148 GLU-180 |
| 2 | ASCL1 | Laevulinic acid | 2098 | 2 | GLY-81 ARG-131 |
| 3 | ASCL1 | Choline | 2160 | 2 | ARG-131 ARG-100 |
| 4 | ASCL1 | Lecithin | 6648 | 9 | GLY-119 ARG-121 ALA-117 LEU-116 ASP-110 ALA-111 GLU-180 ALA-179 SER-144 |
Table 4 (A5): Docking results of ASCL1 receptor with compounds from DANDELION.
| WOOD BETONY | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | GCH1 | Betulinic acid | 4608 | 5 | TRP-53 GLU-56 |
| 2 | GCH1 | Delphinic acid | 2142 | 2 | LYS-54 GLY-55 ARG-57 ALA-120 ILE-121 |
| 3 | GCH1 | Oleonilic acid | 4708 | 1 | HIS-210 |
| 4 | GCH1 | Rosamarinic acid | 4260 | 4 | ILE-113 TYR-175 TYR-109 GLU-56 |
| 5 | GCH1 | Rutin | 5020 | 6 | GLY-55 GLU-56 ILE-113 TYR-175 ARG-178 TYR-109 |
| 6 | GCH1 | Urosolic acid | 4992 | 5 | ARG-178 PRO-58 ARG-57 GLY-55 GLU-56 |
| 7 | GCH1 | Stachydrine | 2524 | 3 | LEU-117 ILE-121 ALA-120 |
Table 4 (B1): Docking results of GCH1 receptor with compounds from WOOD BETONY
| NETTLE | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | GCH1 | Histamine | 2208 | 3 | ASN-118 ALA-120 ILE-121 |
| 2 | GCH1 | Formic acid | 1042 | 1 | LEU-163 |
| 3 | GCH1 | Acetylcholine | 2578 | 1 | THR-112 |
| 4 | GCH1 | Serotonin | 2806 | 0 | |
Table 4(B2): Docking results of GCH1 receptor with compounds from NETTLE.
| PLANT OVAGATO | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | GCH1 | Arabinose | 2158 | 1 | ALA-196 |
| 2 | GCH1 | Xylose | 2182 | 1 | HIS-210 |
| 3 | GCH1 | Valine | 2164 | 1 | GLN-161 |
| 4 | GCH1 | Alanine | 1772 | 1 | ILE-121 |
| 5 | GCH1 | Glutamic acid | 2152 | 3 | ILE-121 ALA-196 GLN-161 |
| 6 | GCH1 | Glycine | 1462 | 1 | GLU-124 |
| 7 | GCH1 | Cysteine | 1930 | 1 | GLN-161 |
| 8 | GCH1 | Lysine | 2490 | 1 | TYR-109 |
| 9 | GCH1 | Leucine | 2410 | 4 | LEU-117 ASN-118 ALA-120 ILE-121 |
| 10 | GCH1 | Tyrosine | 2794 | 4 | ASP-119 ASN-118 ALA-196 ILE-121 |
| 11 | GCH1 | Rhamnose | 2282 | 4 | LEU-117 ASN-118 ALA-120 ILE-121 |
Table 4(B3): Docking results of GCH1 receptor with compounds from PLANTAGO OVATO.
| TURMERIC | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) | No. of interactions | Interacting amino acids |
| 1 | GCH1 | Curcumin | 4332 | 6 | GLY-55 |
| GLU-56 | |||||
| SER-60 | |||||
| PRO-58 | |||||
| ARG-57 | |||||
| ARG-178 | |||||
| 2 | GCH1 | Camphene | 2696 | 0 | |
Table 4(B4): Docking results of GCH1 receptor with compounds from TURMERIC.
| DANDELION | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No.of interactions | Interacting amino acids |
| 1 | GCH1 | Taraxacin | 3368 | 2 | GLU-183 HIS-144 |
| 2 | GCH1 | Laevulinic acid | 2026 | 3 | GLN-161 ALA-196 LEU-117 |
| 3 | GCH1 | Choline | 2128 | 0 | |
| 4 | GCH1 | Lecithin | 6712 | 6 | LEU-165 LEU-163 TYR-156 GLU-124 HIS-126 SER-250 |
Table 4(B5): Docking results of GCH1 receptor with compounds from DANDELION.
| WOOD BETONY | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | MAOB | Betulinic acid | 5148 | 4 | TYR-112 THR-479 GLU-483 ASP-123 |
| 2 | MAOB | Delphinic acid | 2608 | 1 | GLY-13 |
| 3 | MAOB | Oleonilic acid | 5508 | 2 | THR-478 GLU-483 |
| 4 | MAOB | Rosamarinic acid | 5362 | 2 | TYR-393 ARG-36 |
| 5 | MAOB | Rutin | 5034 | 7 | THR-478 ILE-477 ARG-120 ARG-127 ARG-484 ASN-116 THR-480 |
| 6 | MAOB | Urosolic acid | 5442 | 3 | GLU-483 ILE-477 TYR-112 |
| 7 | MAOB | Stachydrine | 2936 | 1 | GLN-206 |
Table 4(C1): Docking results of MAOB receptor with compounds from WOOD BETONY.
| NETTLE | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | MAOB | Histamine | 2674 | 0 | |
| 2 | MAOB | Formic acid | 1360 | 2 | TYR-326 ILE-199 |
| 3 | MAOB | Acetylcholine | 3552 | 0 | |
| 4 | MAOB | Serotonin | 3786 | 2 | TYR-60 SER-59 |
Table 4(C2): Docking results of MAOB receptor with compounds from NETTLE.
| PLANTAGO OVATO | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | MAOB | Arabinose | 2508 | 3 | SER-394 GLY-13 ARG-36 |
| 2 | MAOB | Xylose | 2740 | 2 | GLN-206 TYR-435 |
| 3 | MAOB | Valine | 2682 | 2 | GLU-34 TYR-393 |
| 4 | MAOB | Alanine | 2188 | 1 | VAL-235 |
| 5 | MAOB | Glutamic acid | 2850 | 2 | GLU-34 ALA-263 |
| 6 | MAOB | Glycine | 1798 | 3 | ALA-429 THR-428 ARG-415 |
| 7 | MAOB | Cysteine | 2364 | 1 | VAL-235 |
| 8 | MAOB | Lysine | 3284 | 0 | |
| 9 | MAOB | Leucine | 2822 | 2 | TYR-393 GLU-34 |
| 10 | MAOB | Tyrosine | 3624 | 2 | ARG-42 TYR-393 |
| 11 | MAOB | Rhamnose | 2748 | 2 | TYR-393 ARG-36 |
Table 4(C3): Docking results of MAOB receptor with compounds from PLANTAGO OVATO.
| TURMERIC | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | MAOB | Curcumin | 5900 | 4 | ARG-42 ALA-439 GLU-34 ILE-264 |
| 2 | MAOB | Camphene | 3148 | 1 | TYR-398 |
Table 4(C4): Docking results of MAOB receptor with compounds from TURMERIC.
| DANDELION | |||||
|---|---|---|---|---|---|
| Sl.no | Receptor | Ligand | Docking score (kcal/mol) |
No. of interactions | Interacting amino acids |
| 1 | MAOB | Taraxacin | 4340 | 2 | GLN-206 CYS-172 |
| 2 | MAOB | Laevulinic acid | 2646 | 3 | CYS-172 TYR-435 TYR-188 |
| 3 | MAOB | Choline | 2640 | 0 | |
| 4 | MAOB | Lecithin | 6636 | 5 | ASP-132 GLN-464 SER-465 PRO-467 GLU-466 |
Table 4(C5): Docking results of MAOB receptor with compounds from DANDELION.
Figure 2A: ASCL1 docking images.
Figure 2B: GCH1 docking images.
Figure 2C: MAOB docking images.
ADME
The phytocompounds used in this work are subjected to ADME screening using molinspiration server [16-18]. The results are noted in Table 5 [1]. From the above table it is seen that the compounds betulinic acid, rutin and lecithin have 1, 3 and 1 violations respectively. Compound curcumin successfully clears ADME studies as it shows NO violations in the Lipinski rule of 5.
| ADME Screening | ||||||
|---|---|---|---|---|---|---|
| Ligands | miLogP | TPSA | Natoms | MW | volume | n violations |
| Acetylcholine | -3.56 | 26.3 | 10 | 146.21 | 156.67 | 0 |
| Alanine | -2.69 | 63.32 | 6 | 89.09 | 84.31 | 0 |
| Arabinose ( Oxane-2,3,4,5-tetrol) | -2.22 | 90.15 | 10 | 150.13 | 126.96 | 0 |
| Betulinic acid (Lup-20(29)-en-28-oic acid, 3beta-hydroxy-) | 7.04 | 57.53 | 33 | 456.71 | 472.04 | 1 |
| Camphene | 3.33 | 0 | 10 | 136.24 | 152.37 | 0 |
| Choline | -4.24 | 20.23 | 7 | 104.17 | 120.16 | 0 |
| Curcumin | 2.3 | 93.07 | 27 | 368.38 | 332.18 | 0 |
| Cysteine | -2.71 | 63.32 | 7 | 121.16 | 102.22 | 0 |
| Delphinic acid (Isovaleric acid) | 1.21 | 37.3 | 7 | 102.13 | 106.39 | 0 |
| Formic acid | -0.51 | 37.3 | 3 | 46.02 | 39.64 | 0 |
| Glutamic acid | -3.25 | 100.62 | 10 | 147.13 | 128.36 | 0 |
| Glycine | -2.55 | 63.32 | 5 | 75.07 | 67.73 | 0 |
| Histamine | -0.85 | 54.71 | 8 | 111.15 | 109.77 | 0 |
| Lecithin | 2.69 | 101.97 | 44 | 643.89 | 668.3 | 1 |
| Leucine | -1.38 | 63.32 | 9 | 131.18 | 134.5 | 0 |
| Levulinic acid | -0.35 | 54.37 | 8 | 116.12 | 108.78 | 0 |
| Lysine | -3.18 | 89.34 | 10 | 146.19 | 146.25 | 0 |
| Oleanoic acid | 6.72 | 57.53 | 33 | 456.71 | 471.14 | 1 |
| Rhamnose (6- Methyloxane-2,3,4,5- tetro)l | -1.64 | 90.15 | 11 | 164.16 | 143.55 | 0 |
| Rosmarinic acid (Rosmarinsaure) | 1.63 | 144.52 | 26 | 360.32 | 303.54 | 0 |
| Rutin ( Vitamin P | -1.06 | 269.43 | 43 | 610.52 | 496.07 | 3 |
| Serotonin | 0.57 | 62.04 | 13 | 176.22 | 165.93 | 0 |
| Stachydrine | -5.31 | 40.13 | 10 | 143.19 | 142.62 | 0 |
| Taraxacin | 2.56 | 43.38 | 18 | 242.27 | 220.04 | 0 |
| Tyrosine | -1.71 | 83.55 | 13 | 181.19 | 163.98 | 0 |
| Urosolic acid (Carissic acid) | 6.79 | 57.53 | 33 | 456.71 | 471.49 | 1 |
| Valine | -1.91 | 63.32 | 8 | 117.15 | 117.7 | 0 |
| Xylose (Ribose, D) | -2.22 | 97.98 | 10 | 150.13 | 130.97 | 0 |
Table 5: ADME screening.
Conclusion
As per the virtual screening studies we find the phytocompounds betulinic acid, rutin, lecithin and curcumin docks with all the receptor genes. As per ADME studies compounds betulinic acid, rutin and lecithin cannot be considered as drug lead as they show 1, 3 and 1 violation respectively in ADME studies (as per Table 5). Compound curcumin successfully clears ADME studies (as per Table 5, there are no violations in the ADME properties of curcumin and hence the compound curcumin can be successfully considered as novel drug for phenylketonuria disease.
REFERENCES
- Hegde G, Anuradha M, Bagchi P. Establishing a Remedy for Phenylketonuria Disease from Medicinal Herbs. Intern J Public Mental Health & NeuroSci. 2018;5:20-26.
- Antshel KM. ADHD, Learning and Academic performance in phenylketonuria. Mol GenetMetab. 2010; S1:S52-58.
- Burton BK, Leviton L, Vespa H, Coon H, Longo N, Lundy BD, et al. A diversified approach for PKU treatment: Routine screening yields high incidence of psychiatric distress in phenylketonuria clinics. Mol Genet Metab. 2013;108:8-12.
- Huijbregts SC, de Sonneville LM, Licht R, van Spronsen FJ, Sergeant JA. Short-term dietary interventions in children and adolescents with treated phenylketonuria: effects on neuropsychological outcome of a well-controlled population. J Inherit Metab Dis. 2002;25:419-30.
- Jahja R, van Spronsen FJ, de Sonneville LM, van der Meere JJ, Bosch AM, Hollak CE, et al. Social-cognitive functioning and social skills in patients with early treated phenylketonuria: a PKU-COBESO study. J Inherit Metab Dis. 2016;39:355-362.
- Khalid MS, Ahmed HM. Phenylketonuria: A new look at an old topic, advances in laboratory diagnosis and therapeutic strategies. Int J Health Sci. 2017;11: 63-70.
- Koch R, Burton B, Hoganson G, Peterson R, Rhead W, Rouse B, et al. Phenylketonuria in adulthood: a collaborative study. J Inherit Metab Dis. 2002;25:333-46.
- Moyle JJ, Fox AM, Arthur M, Bynevelt M, Burnett JR. Meta-analysis of Neuropsychological symptoms of adolescents and adults with PKU. Neuropsychol Rev. 2007;17:91-101.
- Naz AH, John C. Phenylketonuria: a review of current and future treatments. Transl Pediatr. 2015;4:304-317.
- Donakonda S, Sinha S, Dighe SN, Rao MRS. System analysis identifies distinct and common functional networks governed by transcription factor ASCL1, in glioma and small cell lung cancer. Mol Biosyst. 2017;13:1481-1494.
- Garavaglia B, Invernizzi F, Carbone ML, Viscardi V, Saracino F, Ghezzi D, et al. GTP-cyclohydrolase I gene mutations in patients with autosomal dominant and recessive GTP-CH1 deficiency: identification and functional characterization of four novel mutations. J Inherit Metab Dis. 2004;27: 455-463.
- Schedin WS, Inoue M, Hromadkova L, Teranishi Y, Yamamoto NG, Wiehager B, et al. Monoamine oxidase B is elevated in Alzheimer disease neurons, is associated with γ-secretase and regulates neuronal amyloid β- peptide levels. Alzheimers Res Ther. 2017;9:57.
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779-815.
- Laskoswki RA, MacArthur MW, Moss DS, Thorton JM. Procheck: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283-291.
- Schneidman DD, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl Acids Res. 2005;33:W363-367.
- Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem. 2000;43:3714-3717.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and Computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 1997;23:4-25.
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, et al. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;S0022-2836(17)30587-9.
Citation: Bagchi P, Visvikis-Sieste S, Kar A (2019) Establishing a Remedy for Phenylketonuria Disease from Indian Ayurvedic Herbs. J Biomol Res Ther 8:177. Doi: 10.35248/2167-7956.19.8.177
Copyright: © 2019 Bagchi P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

