Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2011) Volume 2, Issue 3
Endophytic Bacteria Associated to Sharpshooters (Hemiptera: Cicadellidae), Insect Vectors of Xylella fastidiosa Subsp. pauca
Abstract
Xylella fastidiosa subsp. pauca causes citrus variegated chlorosis (CVC) disease in Brazil, resulting in significant production losses in the citrus industry. X. fastidiosa subsp. pauca is mainly transmitted by three species of sharpshooters (Hemiptera: Cicadellidae) in Brazil; Dilobopterus costalimai (Young), Acrogonia citrina Marucci & Cavichioli and Oncometopia facialis (Signoret). We identified bacterial communities associated with the heads of surface-sterilized insect vectors of X. fastidiosa subsp. pauca that were collected from CVC affected citrus groves in Brazil. Bacteria were isolated and analyzed by amplified ribosomal DNA restriction analysis (ARDRA) and sequencing, revealing the presence, among the most abundant genera, of the well-known citrus endophytes Methylobacterium spp. and Curtobacterium spp. Specific PCR systems for the detection of these genera indicated high frequencies of presence of these bacteria in sharpshooters. The remaining bacterial community was compared in distinct vector species and at different period of the year by denaturing gradient gel electrophoresis (DGGE), showing its responsiveness to the climate change over the year. These results represent a new basis for the knowledge about the interaction symbiotic-pathogenic bacteria inside insect vectors and provide a basis for further work on the biocontrol of plant bacteria like X. fastidiosa.
Keywords: citrus variegated chlorosis (CVC), Curtobacterium sp., Methylobacterium sp.
Introduction
Citrus variegated chlorosis (CVC) was first reported in Brazil in 1987 [1,2] and by 2000, the disease affected already 34% of the 200 million sweet orange trees in state of São Paulo. By 2005, the percentage had increased to 43%, and CVC was present in all citrus growing regions of Brazil [3,4]. The disease is caused by the xylem-limited Gram-negative bacterial pathogen, Xylella fastidiosa subsp. pauca [5,6]. Endophytic microorganisms live within plants without causing apparent harm to the host. We have studied the possible use of endophytes as vehicles to control both phytopathogens and insects [7-10]. Endophytes colonize ecological niches similar to that of phytopathogens [11,12]. The co-localization enables the interaction between both, pathogens and endophytes, creating the possibility to endophytes to act as biocontrol agents [13,14,15]. Biocontrol activity of the endophytic bacterial community may be through the synthesis of structural compounds, such as siderophores and extracellular enzymes [16,17], and the induction and expression of general molecular-based plant immunity [16]. Alternatively, they may act by niche competition, preventing pathogens from becoming established in a host [18]. In citrus, several endophytic bacteria were isolated and Methylobacterium spp. and Curtobacterium flaccumfaciens were further determined as the main endophytic species interacting with X. fastidiosa subsp. pauca [19,20]. These species vary in population density when CVC-affected and asymptomatic plants are compared. Later, Lacava et al. [21] reported that the growth of X. fastidiosa subsp. pauca was inhibited by endophytic C. flaccumfaciens and stimulated by Methylobacterium sp. A similar effect was demonstrated by the reduced severity of the X. fastidiosa subsp. pauca colonization in plants priory colonized by C. flaccumfaciens [22]. Cicadellinae leafhoppers, commonly named sharpshooters, are xylem-feeder insects [23]. An association has been observed between the xylem-feeding habit of sharpshooters and their ability to transmit X. fastidiosa [24-27]. In Brazilian citrus groves, Dilobopterus costalimai, Oncometopia facialis and Acrogonia citrina are the most common sharpshooters found [25-27]. After acquisition of X. fastidiosa by the insects, colonies of bacterial cells were visible in the cibarium and pre-cibarium of transmitting insects attached to the foregut walls [21,28] and also in insect honeydew [29]. The transmission efficiency of bacteria is a measure of the ability to successfully acquire bacteria from an infected plant and transmit to healthy ones. The transmission rates for the main species associated with CVC vary from 1% to 5% [26,30,31]. Many aspects can influence the transmission of a pathogen by an insect vector such as the low concentration of X. fastidiosa subsp. pauca cells in the citrus plants [22] and the low number of colonized vessels in infected plants [32]. The interaction between different bacteria inside the insect foregut may also influence the transmission, as once inside the foregut, bacterial interaction, such as competition for nutrients, space and other complex interactions could occur. In this context, the aims of this work were (i) to access the bacterial population associated with the main sharpshooters responsible for the transmission of X. fastidiosa subsp. pauca in citrus; (ii) to evaluate the diversity of heterotrophic bacteria by amplified ribosomal DNA restriction analysis (ARDRA); (iii) to compare the bacterial community presents in the insect heads from distinct species and collected from citrus at distinct period of the year by denaturing gradient gel electrophoresis (DGGE).
Materials and Methods
Sampled vectors of Xylella fastidiosa subsp. pauca
Insects affiliated to the species Oncometopia facialis, Acrogonia citrina and Dilobopterus costalimai were collected from plants of sweet orange (Citrus sinensis L. Osbeck), cultivated in an orange groves affected by X. fastidiosa subsp. pauca located in the city of Bebedouro (State of São Paulo, Brazil/20°56’58”S48°28’45”W). Three distinct sampling were made, at dates of March 23th, May 05th and June 14th. Insect sweep nets were used to capture the vectors, which were further transported to the laboratory [24].
Isolation of heterotrophic bacteria from insects
At each sampling date, ten insects of each species (O. facialis, A. citrina and D. costalimai) were selected for bacterial isolation (total of 90 analyzed insects: 3 species x 3 dates x 10 insects). Vectors were surface sterilized by immersion in 75% ethanol for two min, transferring them to a container with sodium hypochlorite solution (2% available Cl-) for two min, and rinsing them twice in sterile double-distilled water [33]. After sterilization their heads were cut off, and triturated separately in 1 ml of saline solution (NaCl 0.85 %). An aliquot of 100 μl of this suspension and 1:10 dilutions were plated on non-selective TSB 5% medium (Tryptone Soy Broth, DIFCO) amended with agar (1.5%). Plates were incubated at 28°C for 10 days, colonies were counted, classified based on their morphology and the number of colony forming unities per insect head (CFU/insect head) was determined.
DNA extraction from bacterial isolates
DNA was extracted from fresh bacteria cultures of 120 isolates (Insect Associated Bacterium - IAB) with the following protocol according [24]. A 1.5 ml sample of an overnight bacterial culture was centrifuged for 2 min at 12.000 Χ g and resuspended in 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), recentrifuged, and resuspended in 500 μl of TE buffer plus 0.5 g of 0.1 mm diameter glass beads and 30 μl of 10% sodium dodecyl sulfate. The cells were homogenized for 30 seg in a bead beater (Braun cell homogenizer; B. Braun, Melsungen, Germany). A 500 μl volume of Tris-buffered phenol was then added; the solution was mixed and centrifuged for 10 min at 12.000 Χ g. The aqueous phase was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1), and the DNA was precipitated with isopropanol (1/6 actual volume) incubated for 5 min at room temperature and collected by centrifugation (10 min at 12.000 Χ g). The pellet was washed with 70% ethanol, air dried, and resuspended in 30 μl of double-distilled water.
PCR for 16S rDNA amplification
The amplification of the 16S rDNA from the isolates was carried out in a 50μl reaction volume containing 1μl bacterial DNA (approx. 20 ng), 0.4 μmol-1of each primer (R1378 and PO27F, details in Table 1), 200 μmol-1 of each dNTPs, 3.75 mmol-1 of MgCl2, 2.5 U Taq DNA polymerase (Invitrogen, São Paulo, Brazil) in 10 mmol-1 Tris–HCl (pH 8.3) and 10 mmol-1 KCl. The amplification protocol consisted of an initial step at 94°C for 4 min, followed by 30 amplification cycles of 94°C for 1 min, 62°C for 1 min and 72°C for 1 min with a final extension at 72°C for 10 min. Amplification products were separated by electrophoresis by spotting 5μl of the PCR reaction mixture onto 1% agarose gel and visualized under ultra-violet light after staining with ethidium bromide (0.5 μg ml-1). ARDRA and ribotype selection After the amplification of bacterial 16S rDNA from 120 bacterial isolates (40 isolates from each insect species) the amplified fragments (1360bp) were digested with the restriction enzyme AluI (AG/CT) (Fermentas, Vilnius, Lithuania) for 5 hours at 37°C and visualized in a 1.4% agarose gel stained with ethidium bromide (0.5 μg ml-1) under UV light. The resulting patterns were analyzed, and distinct cleavage patterns were considered as ribotypes [34].
Sequencing of the 16S rDNA gene from the selected isolates
Later, isolate representatives of main ribotypes had their 16S rDNA genes partially sequenced and blasted against the nr/nt database at the GenBank (http://www.ncbi.nlm.nih.gov/BLAST) and classified at Ribosomal Data Project (http://rdp.cme.msu.edu/). Also, the phylogeny of obtained sequences was reconstructed in a tree composed by Insect Associated Bacteria 16S rDNA sequences, sequences from type strains of the genera Methylobacterium and Curtobacterium (obtained from Ribosomal Data Project database), and sequences from previously isolated citrus endophytic bacteria from these genera [17]. Alignment and clustering were performed by Kimura-2 parameter and Neighbour-joinning methodology. The phylogenetic and molecular analyses were conducted using MEGA version 4.1 [35], where 1000 replication of bootstrap analyses were performed.
DNA extraction from insect heads
After surface sterilization the heads of insects were cut off, frozen with liquid nitrogen and triturated, separately, in 1 ml of extraction buffer (10 mM Tris-HCl, 1mM EDTA, pH 8.0, SDS 10%). The total DNA extraction was carried out according to the bacterial DNA extraction protocol [33]. From each insect species, DNA from 15 heads was extracted at each sampling date.
Detection of Methylobacterium mesophilicum and Curtobacterium flaccumfaciens by specific PCR
DNA extracted from insect heads was tested for the presence of M. mesophilicum and C. flaccumfaciens, the most important endophytes associated with X. fastidiosa subsp. pauca, by using specific primers [36,37]. Due to a limitation in the amount of bacterial DNA from one single insect head, a nested approach was applied. First, universal bacterial 16S rDNA PCR amplification, using primers PO27F and R1378 (Table 1), was preformed as described below. Second, two microliters of the first reaction was re-amplified in a nested PCR performed with primers MMV6 and R1378 (Table 1), which generated a product of 300bp when DNA from M. mesophilicum is present. Similarly, the detection of C. flaccumfaciens was made with primers CF16S and 530R (Table 1), which generated a product of 400bp. The PCR reactions consisted of an initial denaturation step of 94°C for 7 min, followed by 25 cycles of 94°C for 1 min, 56°C and 62°C (for M. mesophilicum and C. flaccumfaciens respectively) for 1 min, and 72°C for 1 min and a final extension of 10 min at 72°C. The percentage of positive results was determined.
| Primer | Target DNA | Sequence (5’- 3’) | Reference |
|---|---|---|---|
| MMV6 | Methylobacterium mesophilicum 16SrDNA | ACGTGGAGAGATTCACGG | Rossetto et al., 2000 |
| CF16S | Curtobacterium flaccumfaciens 16SrDNA | ATCAGGAGCTTGCTCCTGTGA | Rossetto et al., 2000 |
| 530R | 16S rDNA | CCGCGGCKGCTGGCAC | Lane, 1985 |
| R1378 | Bacterial 16S rDNA | CGGTGTGTACAAGGCCCGGGAACG | Heuer et al., 1997 |
| U968-GC | Bacterial 16S rDNA | CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG-AACGCGAAGAACCTTAC | Heuer et al., 1997 |
| PO27F | Bacterial 16S rDNA | GAGAGTTTGATCCTGGCTCAG | Lane, 1985 |
Table 1: Primers used in this study, target organism and sequence.
Assessment of the bacterial community in sharpshooter heads by DGGE
The total DNA extracted from insect heads was subjected for DGGE analysis. A nested approach was applied as previously described for specific PCR procedures. First amplification was performed using primers PO27F and R1378 (Table 1). The second amplification was made with DGGE primers R1378 and U968GC (Table 1) as previously described [38]. A PCR mixture was made up of 1 μl of extracted DNA (approx. 5 ng) from the insect heads, 5 μl of 10X Stoffel buffer (10 mM Tris-HCl [pH 8.3], 10 mM KCl), 20 pmol of each primer (Table 1), 200 μmol of each deoxynucleoside triphosphate, 3.75 mM MgCl2, 0.5 μl of formamide and 2.5 U of Taq DNA polymerase Invitrogen (Carlsbad, CA, USA) in a 50 μl final volume. For the amplification protocol, denaturation was carried out at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 2 min, and a final extension of 10 min at 72°C. Five micro liters of the PCR product was analyzed by electrophoresis in a 1.4% agarose gel with 0.5X TBE buffer before running the DGGE gel.
DGGE was performed as previously described [33,39] using the INGENY phor U2 apparatus (INGENY, Goes, The Netherlands). PCR samples were loaded onto 6.0% polyacrylamide gels in 0.5X TAE buffer (20 mM Tris-acetate, 0.5 mM EDTA, pH 7.4). The polyacrylamide gels were made with denaturing gradients ranging from 45 to 65% (where the 100% denaturant contained 7 M urea and 40% formamide). The gels were run for 16 h at 100 V and 60°C, after which the gels were soaked for 30 min in SYBR Green I nucleic acid stain (1:10,000 dilution; Molecular Probes, Leiden, The Netherlands) and immediately photographed under UV light.
In order to compare the DGGE fingerprinting, gels were submitted to analysis by the software GelcomparII (Applied Maths, Kortrigk, Belgium), where images were converted, normalized and band patterns were compared. Cluster analysis of DGGE patterns was performed using the UPGMA method (unweighted pair group method with arithmetic mean) based on the similarity calculated by densitometric Pearson correlation.
Results
Isolation of cultivable bacteria from insects
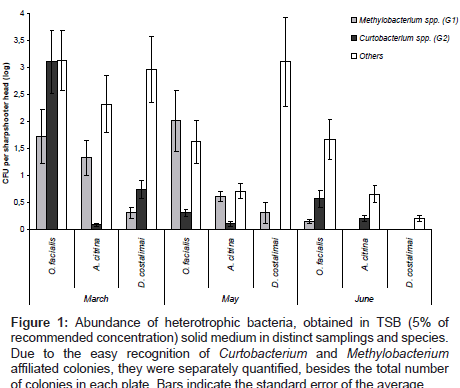
A total of 17.230 bacterial colonies were counted in all isolations from different species and at distinct sampling time. These colonies were classified into three distinct morphological groups; G1) pinkpigmented colonies which appear on plates after seven days of incubations, typically Methylobacterium spp.; G2) dark pink or orange fast grower colonies, typically Curtobacterium spp.; G3) other colonies. The concentration of bacteria (CFU/insect head) belonging to each morphological group is represented in (Figure 1).
Figure 1: Abundance of heterotrophic bacteria, obtained in TSB (5% of recommended concentration) solid medium in distinct samplings and species. Due to the easy recognition of Curtobacterium and Methylobacterium affiliated colonies, they were separately quantified, besides the total number of colonies in each plate. Bars indicate the standard error of the average.
The results show a differential composition of the heterotrophic bacterial communities according to the sharpshooter species and depending on the month of sampling. Abundance of total bacteria was higher from samples taken in March compared to numbers from May and June. Also specific groups varied in frequency along the period of samplings. The populations of group G1 (Methylobacterium) remained constant for O. facialis and D. costalimai in March and May, decreasing in June. From A. citrina the presence of G1 bacteria decreased from May. Group 2 (Curtobacterium) was higher in March for O. facialis and D. costalimai, decreased in May and was undetected in D. costalimai in June. The population of Curtbacterium spp. in A. citrina remained constant during the period of analysis (Figure 1).
ARDRA and sequencing
A subsample of the total number of colonies obtained (120 colonies, 40 isolated from each insect species) was subjected to the genotypic characterization by ARDRA. In total, 16 cleavage patterns were observed, determining the ribotypes constituting the heterotrophic bacterial communities from sharpshooter heads. Among these ribotypes, the colonies from the two targeted groups, Curtobacterium spp and Methylobacterium spp., has revealed a fidelity in the pattern of the 16S rDNA gene cleavage, allowing this approach to measure the proportion of these bacteria within the sampled colonies (Figure 2). Other bacterial haplotypes were characterized randomly and sequences have shown the affiliation of the isolates to the genera Bacillus, Brachybacterium, Brevibacillus, Brevundimonas, Nocardia, Paenibacillus, Pseudoclavibacter, Rhodococcus, Sphingomonas and Staphylococcus (Table 2).
| Sharp shooter species | Ribo type | Isolate | GenBank accession numbers | Best Blast match (similarity) | RDP Classification (taxonomic level) |
|---|---|---|---|---|---|
| O. facialis | I | IAB68 | GU573523 | Methylobacterium radiotolerans strain P (99%) | Methylobacterium radiotolerans AY616142 (97%) |
| IAB47 | GU573507 | Methylobacterium sp. P11E (94%) | Methylobacterium sp. D32231 (86%) | ||
| IAB73 | GU573526 | Methylobacterium sp. PB183 (97%) | Methylobacterium sp. D32231 (96%) | ||
| IAB35 | GU573498 | Methylobacterium sp. C1FA3 (98%) | Uncultured Methylobacteriaceae AY360526 (95%) | ||
| IAB36 | GU573499 | Methylobacterium sp. C1FA3 (98%) | Methylobacterium sp. D32231 (97%) | ||
| IAB76 | GU573529 | Methylobacterium sp. C1FA3 (99%) | Methylobacterium sp. D32231 (98%) | ||
| IAB77 | GU573530 | Methylobacterium sp. C1FA3 (99%) | Methylobacterium sp. D32231 (100%) | ||
| IAB90 | GU573538 | M. radiotolerans strain P (98%) | M. radiotolerans AY616142 (97%) | ||
| II | IAB69 | GU573524 | C.luteum 1P10AnD (98%) | C. luteum X77437 (92%) | |
| IAB74 | GU573527 | C. flaccumfaciens SMW-8 (99%) | C. luteum X77437 (97%) | ||
| IAB34 | GU573497 | C. luteum 1P10AnD (96%) | Curtobacterium sp. AB046364 (87%) | ||
| IAB75 | GU573528 | C. flaccumfaciens pv. basellae (97%) | Curtobacterium sp. AB046364 (91%) | ||
| IAB79 | GU573531 | C. citreum LCR27 (98%) | Curtobacterium sp. AY688357 (97%) | ||
| IAB80 | GU573532 | Curtobacterium sp. LCR25 (98%) | Curtobacterium sp. AB046364 (92%) | ||
| IAB41 | GU573502 | C. flaccumfaciens SMW-8 (97%) | C. luteum X77437 (93%) | ||
| IAB42 | GU573503 | C citreum LCR27 (97%) | Curtobacterium sp. AY688357 (91%) | ||
| IAB83 | GU573534 | C. luteum 1P10AnD(98%) | Curtobacterium sp. AB046364 (93%) | ||
| IAB70 | GU573525 | C.luteum 1P10AnD (98%) | Curtobacterium sp. AB046364 (94%) | ||
| IAB33 | GU573496 | C. luteum X77437 (97%) | C. luteum X77437 (93%) | ||
| Others | IAB91 | GU57353 | Brevibacillus brevis NBRC 100599 (98%) | Brevibacillus agri AB112716 (95%) | |
| IAB48 | GU573508 | Sphingomonas phyllosphaerae FA1 (97%) | Sphingomonas phyllosphaerae AY453855 (94%) | ||
| A. citrina | II | IAB63 | GU573520 | C.luteum 1P10AnD (98%) | C. flaccumfaciens EF063717 (92%) |
| IAB85 | GU573535 | C.luteum 1P10AnD (97%) | Curtobacterium sp. AB046364 (95%) | ||
| Others | IAB53 | GU573513 | Bacillus bataviensis PBA(98%) | Bacillus sp. DQ275174 (93%) | |
| IAB56 | GU573515 | B. megaterium DQ447778 (98%) | Bacillus sp. EU276114 (96%) | ||
| IAB58 | GU573516 | Brevundimonas sp SB4 (97%) | Brevundimonas sp. DQ981459 (94%) | ||
| IAB46 | GU573506 | B. firmus SK20 (97%) | Bacillus firmus AJ717384 (93%) | ||
| IAB87 | GU573537 | Brachybacterium paraconglomeratum 13636D (95%) | Brachybacterium paraconglomeratum AB098579 (89%) | ||
| IAB86 | GU573536 | Brachybacterium phenoliresistens phenol-A (99%) | Brachybacterium sp. EU249983 (96%) | ||
| IAB62 | GU573519 | Staphylococcus pasteuri BCCS 018 (99%) | Staphylococcus sp. AJ276810 (95%) | ||
| IAB59 | GU573517 | Pseudoclavibacter helvolus BH16SR (97%) | Pseudoclavibacter helvolus X77440 (88%) | ||
| D. costalimai | Others | IAB50 | GU573510 | Nocardia corynebacterioides NRRL 21057 (97%) | Nocardia beijingensis AB094641 (91%) |
| IAB51 | GU573511 | Rhodococcus sp. lxb-6(96%) | Rhodococcus sp. AJ002093 (89%) | ||
| IAB45 | GU573505 | Bacillus firmus UST981101-007 (98%) | Bacillus firmus AJ717383 (95%) | ||
| IAB81 | GU573533 | Paenibacillus ginsengagri (97%) | Paenibacillus sp. AF395029 (96%) |
Table 2: Identification of distinct ribotypes obtained from the cleavage patterns of the16S rDNA gene from bacterial isolates from sharpshooter (Insect Associated Bacteria). Sequences classifications were made by closest Blast match and by classification at Ribosomal Data Project.
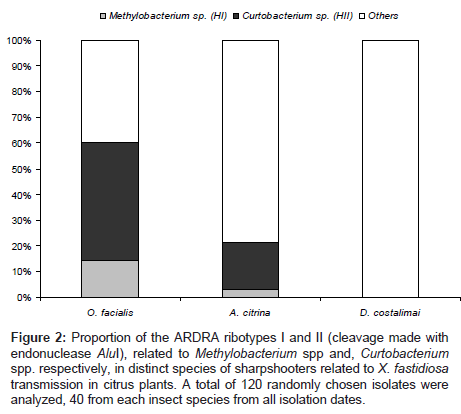
Figure 2: Proportion of the ARDRA ribotypes I and II (cleavage made with endonuclease AluI), related to Methylobacterium spp and, Curtobacterium spp. respectively, in distinct species of sharpshooters related to X. fastidiosa transmission in citrus plants. A total of 120 randomly chosen isolates were analyzed, 40 from each insect species from all isolation dates.
Correlating the number of ribotypes found in colonies from each insect species, we observed a higher occurrence of the ribotype II (Curtobacterium spp.) in species O. facialis and A. citrina, and a similar trend but in lower number for the occurrence of Methylobacterium spp. (ribotype I) (Figure 2).
Confirmation of the Curtobacterium spp and Methylobacterium spp. colonization by specific PCR
DNA samples directly extracted from insect heads were used for specific amplification of DNA from bacteria affiliated with the genera Curtobacterium and Methylobacterium. DNA from 15 heads of each insect species was extracted at each sampling date. The number of positive samples was unique to each sharpshooter species. The detection of DNA from Curtobacterium spp. was positive in 89.6% of the samples from O. facialis, 39.1% of D. costalimai and 70% of A. citrina. In a similar analysis for Methylobacterium spp. the numbers of positive PCR were 51.7% for O. facialis, 8.7% for D. costalimai and 20% of the A. citrina. The results from isolation and ARDRA ribotyping are presented in comparison to previous work in the (Table 3).
| Analyzed parameters | Sharpshooter species | ||
|---|---|---|---|
| O. facialis | A. citrina | D. costalimai | |
| Frequency of Methylobacterium spp. isolation (G1)1 | 47 | 8 | 60.4 |
| Frequency of Curtobacterium spp. isolation (G2)1 | 450 | 0.36 | 1.6 |
| Percentage ofpositive specific PCR for Methylobacterium spp.2 | 51.7% | 20.0% | 8.7% |
| Percentage ofpositive specific PCR for Curtobacterium spp.2 | 89.6% | 70.0% | 39.1% |
| Number of isolates with ribotype I (Methylobacterium spp.) (out of 120) | 10 | 1 | 0 |
| Number of isolates with ribotype II (Curtobacterium spp.) (out of 120) | 32 | 6 | 0 |
| Transmission efficiency of Xfp (according to Krügner et al., 2000) |
1% | 2% | 5% |
1Average of the number of bacteria isolated from each insect species in all sampling dates. CFU/head.
2Percentage of positive specific PCR for the detection of Curtobacterium or Methylobacterium using specific primers in a nested PCR. To each insect species 15 individuals were tested from each sampling date resulting in 45 insects tested.
Table 3: A comparative analysis of the Curtobacterium spp. and Methylobacterium spp. isolation and the efficiency of the sharpshooter species in transmitting Xylella fastidiosa subsp. pauca (Xfp), previously determined (Krügner et al., 2000).
Phylogeny of sharpshooter isolates and similarity to citrusendophytic bacteria
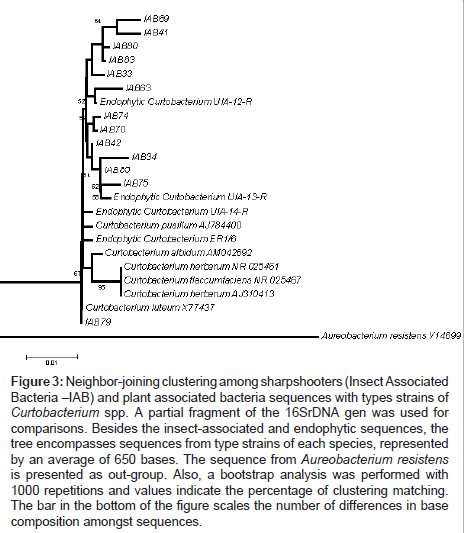
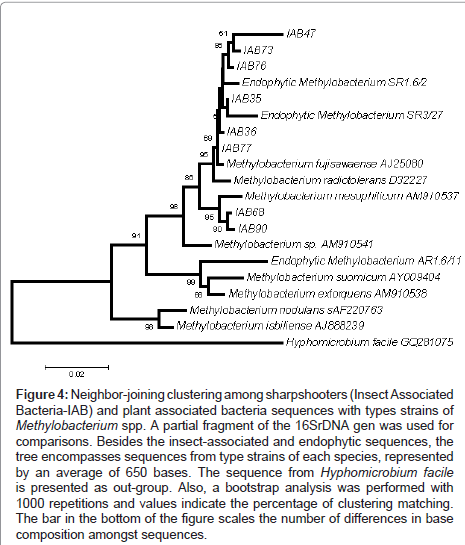
Another point found in this work was the comparative phylogeny of isolates from both targeted groups (Insect Associated Bacteria - IAB) with endophytic isolates obtained in previous works. UIA- 12-R, UIA-13-R, UIA-14-R and ER1.6 are16S rDNA sequences from Curtobacterium flaccumfaciens and AR1.6/11, SR1.6/2 and SR3/27 are from Methylobacterium species previously isolated from citrus [17].One can observe the clustering of IAB isolated in this work with endophytic strains isolated previously from citrus trees, more specifically C. flaccumfaciens strain UIA-12-R (Figure 3) and Methylobacterium strains SR1.6/2 and SR3/27 (Figure 4). In general, bacteria isolated in this study clustered together.
Figure 3: Neighbor-joining clustering among sharpshooters (Insect Associated Bacteria –IAB) and plant associated bacteria sequences with types strains of Curtobacterium spp. A partial fragment of the 16SrDNA gen was used for comparisons. Besides the insect-associated and endophytic sequences, the tree encompasses sequences from type strains of each species, represented by an average of 650 bases. The sequence from Aureobacterium resistens is presented as out-group. Also, a bootstrap analysis was performed with 1000 repetitions and values indicate the percentage of clustering matching. The bar in the bottom of the figure scales the number of differences in base composition amongst sequences.
Figure 4: Neighbor-joining clustering among sharpshooters (Insect Associated Bacteria-IAB) and plant associated bacteria sequences with types strains of Methylobacterium spp. A partial fragment of the 16SrDNA gen was used for comparisons. Besides the insect-associated and endophytic sequences, the tree encompasses sequences from type strains of each species, represented by an average of 650 bases. The sequence from Hyphomicrobium facile is presented as out-group. Also, a bootstrap analysis was performed with 1000 repetitions and values indicate the percentage of clustering matching. The bar in the bottom of the figure scales the number of differences in base composition amongst sequences.
Analysis of the bacterial communities of sharpshooter heads by DGGE
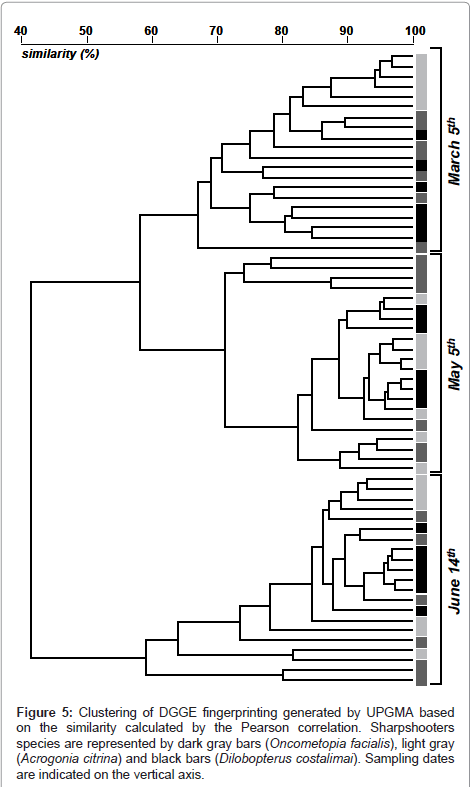
The DGGE analysis showed variable fingerprinting according to the period when insects were collected. The first separation was observed for samples collected in June, with a further sub-branching for samples from May and March (Figure 5). These variations were based on changes in the abundance of bands and also in the intensity of similar bands found in distinct samples. Also, within each major branching it is possible to separate the DGGE pattern according to the vector species, with a more deterministic separation for the species A. citrina (Figure 5). For other species, D. costalimai and O. facialis, the species-selection was lower, as evidenced by the mixing of samples from these species of sharpshooters.
Figure 5: Clustering of DGGE fingerprinting generated by UPGMA based on the similarity calculated by the Pearson correlation. Sharpshooters species are represented by dark gray bars (Oncometopia facialis), light gray (Acrogonia citrina) and black bars (Dilobopterus costalimai). Sampling dates are indicated on the vertical axis.
Discussion
The role of associated microbial community in the pathogenic potential of X. fastidiosa has been discussed in the literature [12,14,17,18,37], where authors state that these microorganisms can partially inhibit or promote the growth of X. fastidiosa inside the citrus plants. However, there was a lack of knowledge about the occurrence and frequency of endophytic bacteria commonly found inside the insect vectors, which transmit pathogens from plant-to-plant. In order to fill this gap this study was conducted, using three insect species described as X. fastidiosa vectors, generating the first insights in this important aspect, allowing further focused studies on the biological control or symbiotic control of this pathogen in citrus groves. We focused our approaches in detecting and quantifying two major citrus endophytes in the selected insects. However, a culture-independent approach was also used to describe changes in bacterial communities associated with these vectors.
Differences in bacterial communities associated with insects suggest complex interactions between insects, host plants, bacteria and environment. In March, when the temperatures were around 25°C and the precipitation 160 mm the bacterial community was denser with a higher number of bacterial groups, when compared to the months of May and June when the temperatures and the precipitation were lower (20°C, 15°C and 80 mm, 60mm respectively). Each insect contained an associated bacterial community that was altered in time and in different environmental conditions. Climate shifts affect not only the bacterial community in plants but also the behavior of insects [31], which choose alternative host plants during different periods of the year, and host plant conditions (citrus or adjacent ones), which change depending on temperature and precipitation on the area.
The DGGE technique has been used to study bacterial communities [39]. Reeson et al. [40] described the bacterial communities associated with wasps (Vespula germanica) by applying DGGE-based methods as well as Lacava et al. [33] who used DGGE to characterize microbe in glassy-winged sharpshooter (GWSS), the main insect vector of X. fastidiosa subsp. piercei [6] causing Pierce’s disease (PD) in the USA [22]. Lacava et al. [33] found M. extorquens and C. flaccumfaciens as part of the bacterial community of GWSS in the USA. In addition, Kirkpatrick and Wilhelm [41] have also isolated strains of C. flaccumfaciens as part of endophytic bacterial community of grapevine in California, USA. Also, C. flaccumfaciens was consistently isolated as an endophytic bacterium from citrus plants in Brazil [17,18].
In the present study, Curtobacterium sp. was the most abundant bacteria colonizing insect heads. Curtobacterium flaccumfaciens was implicated in playing an important role in the prevention of CVC symptoms in citrus trees [17,18,33]. The presence of the citrus endophyte, Curtobacterium sp., colonizing the insect heads could explain why the transmission efficiency of X. fastidiosa subsp. pauca by vectors is low (5 to 10%), when compared to the transmission of X. fastidiosa subsp. fastidiosa by GWSS, which cause Pierece’s disease in grapevines (45%) [26,31]. Table 3 illustrates that the genus Curtobacterium, can play an important role in the transmission of X. fastidiosa subsp. pauca. The presence of this Curtobacterium has been cited in several works where the interaction of these bacteria with X.fastidiosa had been studied. High population of Curtobacterium were observed in asymptomatic citrus plant [19,20], A less severe colonization of plants by X.fastidiosa was observed in plants previously colonized by C.flaccumfaciens [22] and in this study, a high population of Curtobacterium was observed associated to insects with lower transmission rate of X.fastidiosa.These observations show a colonization pattern what suggest an interaction between both species where Curtobacterium would affect negatively the growth of X.fastidiosa and so the transmission and development of CVC symptoms. Curtobacterium could be influencing pathogen adhesion to the vector foregut or inhibiting growth of the pathogen.
The bacterial communities associated with vector insects and plants differ in abundance through the yearly season. Endophytic bacteria could influence disease development by reducing the insect transmission efficiency due to competition with pathogens in host plants and also in insect foreguts. In addition the bacterial communities in the foregut of insect vectors of X. fastidiosa subs p. pauca changed with time, environmental conditions and in different insect species. However, members of the genus Curtobacterium were consistently detected in the sharpshooters foregut and are commonly isolated from the xylem of citrus plants [17], and because of this, they may be candidates for biological control.
In a recent review, Lacava et al. [15] suggested C. flaccunfaciens is a potential candidate for biological control of CVC because an antagonism between C. flaccumfaciens and X. fastidiosa subsp. pauca was strongly indicated in vitro and in vivo [17,18,33] including inhibition of growth of X. fastidiosa subsp. pauca and reduced severity of the disease symptoms in the presence of this phytopathogen. The ability demonstrated by C. flaccumfaciens to colonize plant tissues in the presence of X. fastidiosa subsp. pauca and the reduction of disease symptoms caused by X. fastidiosa subsp. pauca [19] are prerequisites for the use of this endophytic bacterium as a biocontrol agent. Since members of the genus Curtobacterium were consistently detected in Acrogonia citrina Marucci & Cavichioli and Oncometopia facialis (Signoret), two of the main insect vectors of X. fastidiosa subs p. pauca as demonstrated in the present study, they fulfill another requirement of candidates for biological control of X. fastidiosa subsp. pauca [19], i.e. they can colonize both the insect vectors of X. fastidiosa subsp. pauca and citrus plants.
Acknowledgements
This work was supported by a grant from FAPESP (06/55494-4). We thank CNPq for the fellowship to C.S.Gai. Also, we thank the Citriculture Centre in Cordeirópolis, SP, Brazil and the Transmission Laboratory (ESALQ/USP) for the field collection of the insects
References
- Rossetti V, De Negri D (1990) Citrus Variegated Chlorosis (CVC): Review. Orange. 11: 1-14.
- Lee RF, Kerrick KS, Beretta MJG, Chagas CM, Rossetti V (1991). Citrus variegated chlorosis: a new destructive disease of citrus in Brazil. Citrus Ind 72: 12-13.
- Bové JM, Ayres AJ (2007) Etiology of three recent diseases of citrus in São Paulo State: Sudden death, variegated chlorosis and huanglongbing. IUBMB Life 59: 346-354.
- Tennant PF, Robinson D, Fisher L, Bennett SM, Hutton D, et al. (2009) Diseases and pests of citrus (Citrus spp.). Tree For Sci Biotech 3: 81–107.
- Hartung JS, Beretta J, Brlansky RH, Spisso J, Lee RF (1994) Citrus variegated chlorosis bacterium: axenic culture, pathogenicity, and serological relationships with other strains of Xylella fastidiosa. Phytopathology 84: 591-597.
- Schaad NW, Postnikova E, Lacy G, Fatmi M, Chang CJ (2004) Xylella fastidiosa subspecies: X. fastidiosa subsp. piercei, subsp. nov., X. fastidiosa subsp. multiplex, subsp. nov., X. fastidiosa subsp. pauca, subsp. nov. Syst Appl Microbiol 27: 290-300.
- Azevedo JL, Maccheroni W, Pereira JO, Araújo WL (2000) Endophytic microorganisms: A review on insect control and recent advances on tropical plants. Electronic J of Biotechnol 3.
- Kozdrój J, Trevors JT, Elsas JDV (2004) Influence of introduced potential biocontrol agents on maize seedling growth and bacterial community structure in the rhizosphere. Soil Biol Biochem 36: 1775-1784.
- Kuklinsky-Sobral J, Araújo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, et al. (2004) Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Env Microbiol 6: 1244-1251.
- Kavino M, Harish S, Kumar N, Saravanakumar D, Damodaran T, et al.(2007) Rhizosphere and endophytic bacteria for induction of systemic resistance of banana plantlets against bunchy top virus. Soil Biol Biochem 39: 1087-1098.
- Lacava PT, Li WB, Araújo WL, Azevedo JL, Hartung JS (2006) Rapid, specific and quantitative assays for the detection of the endophytic bacterium Methylobacterium mesophilicum in plants. J Microbiol Meth 65: 535-541.
- Gai CS, Lacava PT, Maccheroni W, Glienke C, Araújo WL, et al. (2009a) Diversity of endophytic yeasts from sweet orange and their localization by scanning electron microscopy. J Basic Microbiol 49: 441-451.
- Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW (1997) Bacterial endophytes in agricultural crops. Can J Microbiol 43: 895-914.
- Gai CS, Lacava PT, Quecine MC, Auriac MC, Lopes JRS, et al. (2009b) Transmission of Methylobacterium mesophilicum by Bucephalogonia xanthophis for paratransgenic control strategy of Citrus Variegated Chlorosis. J Microbiol 47: 448-454.
- Lacava PT, Azevedo JL, Miller TA, Hartung JS (2009) Interaction of Xylella fastidiosa and endophytic bacteria in citrus: a review. Tree For Sci Biotech 3: 40-48.
- Araújo WL, Maccheroni W, Aguilar-Vildoso CI, Barroso PA, Saridakis HO, et al. (2001) Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can J Microbiol 47: 229-236.
- Araújo WL, Marcon J, Maccheroni W Jr, Elsas JDV, Vuurde JWV, et al. (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in Citrus Plants. Appl Environ Microbiol 68: 4906-4914.
- Lacava PT, Araújo WL, Marcon J, Maccheroni W Jr, Azevedo JL (2004) Interaction between endophytic bacteria from citrus plants and the phytopathogenic bacteria Xylella fastidiosa , causal agent of citrus-variegated chlorosis. Lett Appl Microbiol 39: 55-59.
- Lacava PT, Li WB, Araújo WL, Azevedo JL, Hartung JS (2007a) The endophyte Curtobacterium flaccumfaciens reduces symptoms caused by Xylella fastidiosa in Catharanthus roseus. J Microbiol 45: 388–393.
- Young DA (1968) Taxonomic Study of the Cicadellinae (Homoptera: Cicadellidae). Part 1. Proconiini. Bull. United States Nat Mus 261: 1- 287.
- Purcell AH, Finlay AH (1979) Evidence for noncirculative transmission of Pierce’s disease bacterium by sharshooter leafhoppers. Phytopathology 69: 393-395.
- Almeida RP, Purcell AH (2003) Transmission of Xylella fastidiosa to grapevines by Homalodisca coagulata (Hemiptera: Cicadellidae). J Econ Entomol 96: 264- 271.
- Costa HS, Blua MS, Bethke JA, Redak RA (2000) Transmission of Xylella fastidiosa to oleander by the glasswinged sharpshooter, Homalodisca coagulata. HortScience 35: 1265-1267.
- Ramirez JL, Perring TM, Miller TA (2008) Fate of a genetically modified bacterium in foregut of glassy-winged sharpshooter (Hemiptera: Cicadellidae). J Econ Entomol 101: 1519-1525.
- Lopes JRS, Beretta MJG, Harakava R, Almeida RPP, Krügner R, et al. (1996) Confirmation of transmission by sharpshooters of the causal agent of citrus variegated chlorosis, Xylella fastidiosa . Fitopat Bras 21: 343.
- Krügner R, Lopes MTVC, Santos JS, Beretta MJG, Lopes JRS (2000) Transmission efficiency of Xylella fastidiosa to citrus by sharpshooters and identification of two vector species. In: Conference of the International Organization of Citrus Virologists, Riverside, 423.
- Yamamoto PT, Roberto SR, Pria Júnior WD, Felippe MR, Miranda VS, et al. (2002) Transmission of Xylella fastidiosa by sharpshooters and Homolodisca ignorata Acrogonia virescens (Hemiptera: Cicadellidae) in citrus. Summa Phyt 28: 178-181.
- Newman KL, Almeida RPP, Purcell AH, Lindow SE (2003) Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl Environ Microb 69: 7319–7327.
- Rodrigues JL, Silva-Stenico ME, De Souza AN, Lopes JR, Tsai SM (2006) In situ probing of Xylella fastidiosa in honeydew of a xylem sap-feeding insect using 16S rDNA-targeted fluorescent oligonucleotides. Env Microbiol 8: 747– 754.
- Marucci RC (2003) Efficiency of Xylella fastidiosa transmission by sharpshooters (Hemiptera, Cicadellidae) in Citrus sinensis (L.) Osbeck and Coffea arabica L. Ph.D. thesis, University of São Paulo, Piracicaba. São Paulo.
- Redak RA, Purcell AH, Lopes JR, Blua MJ, Mizell III RF, et al. (2004) The biology of xylem fluid feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu Rev Entomol 49: 243-270.
- Alves E (2003) Interaction of Xylella fastidiosa with different cultivars of Nicotiana tabacum: a comparison of colonization patterns. J Phytopathol 151: 500-506.
- Lacava PT, Parker J, Andreote FD, Dini-Andreote F, Ramirez JL, et al. (2007b) Analysis of the bacterial community in glassy-winged sharpshooter heads. Entomol Res 37: 261–266.
- Assumpção LC, Lacava PT, Dias ACF, Azevedo JL, Menten JOM (2009) Diversity and biotechnological potential of endophytic bacterial community of soybean seeds. Braz J Agr Res 44: 503-510.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599.
- Rossetto PB, Araújo WL, Maccheroni W, Azevedo JL (2000) Development of primer specific for the detection of endophytic bacteria in citrus. Simp Internat 8th scientific initiative of USP, ESALQ / USP. 8th, ESALQ/USP.
- Andreote FD, Lacava PT, Gai CS, Araújo WL, Maccheroni W, et al. (2006) Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa . Can J Microbiol 52: 419-426.
- Heuer H, Krsek M, Baker P, Smalla K, Wellington EM (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rDNA and gel electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63: 3233-3241.
- Muyzer G, Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rDNA. Appl Environ Microb 59: 695-700.
- Reeson AF, Jankovic T, Kasper ML, Rogers S, Austin AD (2003) Application of 16S rDNA-DGGE to examine the microbial ecology associated with a social wasp Vespula germanica. Insect Mol Biol 12: 85-91.
- Kirkpatrick B, Wilhelm M (2007) Evaluation of grapevine endophytic bacteria for control of Pierce’s disease. Pierce’s disease research symposium. San Diego 203-206.
Copyright: © 2011 Gai CS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.