Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Mini Review - (2021) Volume 12, Issue 1
Dynamic Changes in Microbiome Composition and Genomic Functional Potentials in Bovine Mastitis
M. Nazmul Hoque1,2, Munawar Sultana1 and M. Anwar Hossain1,3*2Department of Gynecology, Obstetrics and Reproductive Health, Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur-1706, Bangladesh
3Jashore University of Science and Technology, Jashore-7408, Bangladesh
Received: 22-Dec-2020 Published: 12-Jan-2021, DOI: 10.35248/2153-0602.21.12.232
Abstract
We provide a mini review on the previously published study entitled “Microbiome Dynamics of Bovine Mastitis Progression and Genomic Determinants”, where we reported the possible dynamic changes in microbiome compositions favored by their genomic functional potentials in different pathophysiological states of bovine mastitis. Using the cutting-edge whole metagenome sequencing (WMS) approach, we reported distinct variation in microbiome composition and abundances across the clinical mastitis (CM), recurrent clinical mastitis (RCM), subclinical mastitis (SCM) and healthy (H) milk metagenomes (CM>H>RCM>SCM). Bacteria were the predominating microbial domain (>99.0% relative abundance) followed by archaea and viruses. Dynamic changes in bacteriome composition across the four metagenomes were numerically dominated by 67.19% inclusion of previously unreported opportunistic strains in mastitis metagenomes. This study also reported the unique and shared distribution of microbiomes across these metagenomes. In addition to microbiome composition and diversity, the study reported the association of several virulence factors-associated genes (VFGs), and antibiotic resistant genes (AGRs) in CM, RCM, SCM, and H-microbiomes. Functional annotation detected several metabolic pathways related to different episodes of mastitis. Therefore, the published data revealed that changes in microbiome composition in different types of mastitis, concurrent assessment of VFGS, ARGs, and genomic functional potentials can contribute to develop microbiome-based diagnostics, and therapeutics for mastitis, and carries significant implications on curtailing the economic fallout from this disease
Keywords
Microbiome Composition; Metabolic Functions; Changes; Clinical; Recurrent Clinical and Subclinical Mastitis
Introduction
Mastitis is the inflammation of the mammary gland and/or quarters [1], and represents the foremost disease-driven challenge in milk production faced by the global dairy industry [2]. The milk microbiota composition is an important determinant of mammalian health [3], and, therefore, plays an important role in udder health by interacting with the immune and metabolic functions of the cow [4], the spread of virulence factors, and the spread of antimicrobial resistance genes (ARGs) [4,5]. Previously, we reported that the microbiome signature in bovine CM is associated with functional biases [5], and observed distinct shifts in the composition of microbiomes in CM and H milk. We also reported that resistomes, which are the potential key factor in disease complication and recurrence, could be concurrent to microbiome signature [6]. Although bovine mastitis microbiome comprises bacteria, archaea, viruses and other microorganisms [5,6], until now, most researchers have mainly focused on the bacterial component of the milk microbiomes. Moreover, a plethora of archaeal and viral entities might be present in association with the bacteria, with relevant physiological and pathological implications for their host [2,5-7]. Bovine mastitis microbiomes showed a paradigm shift in taxonomic diversity and composition according to disease states (clinical, subclinical and recurrent) [2]. Following pathogen invasion and subsequent establishment in the mammary gland, either CM or subclinical mastitis (SCM) may present itself. Bovine CM is one of the most frequent diseases affecting the global dairy industry [8], and diverse groups of microbial communities colonizing the mammary gland and/or quarters facilitate their proliferation during the disease process [8]. One of the very frustrating aspects of bovine CM is its recurrent nature which is caused by persistent intra-mammary infections (IMI) [9]. Almost every dairy herd has cows with SCM, and a variety of pathogens can establish chronic infections which may only occasionally manifest the clinical signs of mastitis [9,10]. The mastitis milk microbiome composition was largely dominated by bacteria (of 99.58%), however a fraction of other microbial domains including archaea (0.24%), and viruses (0.18%) are also be detected [2]. The microbial community present within the mammary gland and/or quarters is being increasingly recognized as a key regulator of metabolic and immune homeostasis [5,6], and a mediator of antimicrobial resistance [6]. The resistomes or ARGs, which exist in both pathogenic [11] and nonpathogenic commensal bacteria is frequently carried on mobile genetic elements. Similarly, the virulome, the set of genes encoding virulence, can also be carried on the mobilome [11], and thus, many VFGs easily spread in bacterial populations by horizontal gene transfer, converting mutualistic or commensal bacteria into potential opportunistic pathogens [12,13].
Literature Review
Culture-independent whole metagenome sequencing (WMS) investigation of the bovine milk microbiome revealed the presence and diversity of microbiomes in milk from healthy and infected mammary glands far greater than previously described [5-8]. We investigated that microbial communities are changed across multiple sample categories of the same disease, and in the healthy condition of the mammary glands. Moreover, the overexpression of genes related to ARGs, VFGs, and several biochemical pathways coming from the metabolic activities of the microbiome is a crucial factor for the development and progression of mastitis [2,5,6]. Therefore, the study for the first time reported potential alterations in microbiome composition along with their VFGs, ARGs, and metabolic potentials in different categories of bovine mastitis (CM, RCM, SCM), and healthy milk through cutting-edge WMS approach.
We found that both the number of observed species/strains remained significantly (p=0.003, Kruskal-Wallis test) higher in CM and RCM groups compared to SCM and H groups. We also observed significant differences (p=0.021, Kruskal-Wallis test) in the milk microbial community structure (i.e., beta diversity) among the CM, RCM, SCM and H milk groups. Principal coordinate analysis (PCoA), measured on the Bray-Curtis distance method, and non-metric multidimensional scaling (NMDS) ordination plots, as measured by weighted-UniFrac distance at strain level, showed clear segregation of samples by the experimental groups. The CM microbiomes had close association with RCM microbiomes followed by SCM and H milk microbes. The present findings of higher taxonomic resolution and predicted protein functions, partially consistent with our previous report [5,6], and many of recent 16S rRNA partial gene-based studies [14-16]. The microbiome diversity (alpha and beta) measures provided that microbial dysbiosis is closely linked to different stages of mastitis. Compared to our previous study [5,6], we observed increased microbial diversity and species richness in CM, RCM and SCM-metagenomes than the healthy-controls. Regardless of higher taxonomic abundances, the bovine mastitis-associated microbiome remained inconsistent, and fluctuates more within CM, RCM and SCM-metagenomes than those of H-milk [2,5,6].
Bovine mastitis (clinical, recurrent clinical and subclinical) is predominantly caused by the bacteria of both environmental and commensal (from udder, teat, skin and gut) origin. Under immunosuppression or stress conditions (when the cow suffers from a severe negative energy balance at the onset, in-andaround lactation, and other environmental stress), environmental and commensal bacteria act as potential opportunists to manifest different episodes of mastitis. A plethora of other microbes including archaea and viruses simultaneously follow the same opportunistic pathways as bacteria. However, the possible dynamic shifts in microbiome compositions in different conditions of bovine mastitis are determined by its favoring genomic potentials including virulence, antimicrobial resistance and metabolic functions (please see further details in the original publication [2].
The dynamic changes in bacterial, archaeal and viral fractions of the bovine mastitis (CM, RCM, SCM) and healthy (H) milk microbiomes is shown in Figure 1. The composition of microbial taxa remained much higher in CM milk followed by H, RCM and SCM milk metagenomes. In that investigation, we reported 18 bacterial phyla (16, 11, 4 and 13 in CM, RCM, SCM and H, respectively), and of them 4 phyla were found to be common across the four metagenomes. The associated bacterial phyla were numerically dominated by Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes (comprising >99% of relative abundance) in mastitis and healthy milk metagenomes. Using the state-of-the-art bioinformatics tool (PathoScope 2.0) we reported the strain-level taxonomic profile of bacteria (n=442 bacterial strains), and of the identified strains, 15.44%, 12.30%, 3.75% and 20.14% had unique associations in CM, RCM, SCM, and H milk, respectively. Across these four metagenomes, 67.19% of the detected bacterial strains were previously unreported, and of them, Nocardia pseudobrasiliensis, Aeromonas veronii B565, Chryseobacterium sp. Leaf 405, Pantoea dispersa EGD-AAK13, Serratia marcescens subsp. marcescens Db11, Klebsiella oxytoca KA-2, and Citrobacter freundii CFNIH1 were most prevalent. Many of these microbiotas may act as potential opportunist by interfering with metabolism, host defense, and immune development [5,17,18] to producing different magnitude of udder infections. Our present findings also revealed the potential existence of endogenous entero-mammary pathway, and through this axis the gut or rumen microbiome migrate to the mammary gland, and manifested different episodes of mastitis by using the very efficient strategies of colonizing and invading mammary tissues through adhesion [13], thus damaging host cells and fighting with immune systems [5,6,13,19]. The healthy-milk metagenome was also dominated by different strains of environmental, gut and animal skin originating microbes. Though the pathogenic mechanism of these commensal microbe is largely unknown, however can cause opportunistic infections of the mammary glands and/or quarters with or without varying degree of clinical episodes by producing different virulence factors [20] particularly in immunocompromised hosts [2,5,6,17].
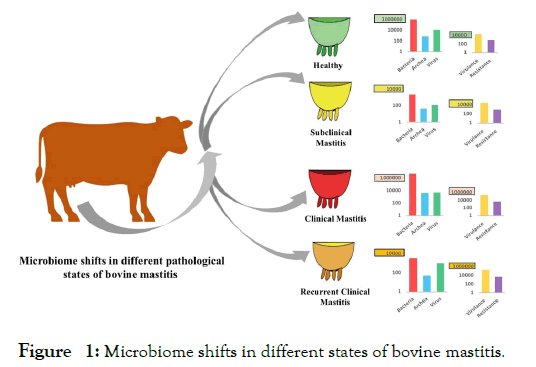
Figure 1: Microbiome shifts in different states of bovine mastitis.
In addition, this study for the first time reported the association of 58 archaeal and 48 viral genera along with bacterial fraction of the microbiomes to causing different episodes of bovine mastitis, and of the detected genera, 12.06% and 20.83% of the archaeal and viral genera, respectively were reported to be common in the samples of four metagenomes (Figure 1). However, unlike bacteria, the diversity and composition of archaea [21] and viruses [22] always remained much lower in both mastitis and healthy milk [2,5,6]. The CM-milk metagenome always had higher taxonomic compositions and abundances of both archaeal and viral fractions followed by H, RCM and SCM-milk metagenomes. Even though, the role of these accompanying microbiotas in the pathophysiology of bovine mastitis has not been understood within the frames of a typical host-pathogen interaction, however, these microbiotas could cease the opportunity during the pathological changes in the mammary glands created by bacteria [17,21]. The study predicted that archaea and virus may occupy the microenvironments suitable for anaerobic metabolism, make way for opportunistic pathogens (pathobionts), already present in milk, and/or gain secondary access to the mammary gland when host is in unnatural condition or immune compromised [23], and follow the similar virulence mechanisms of bacterial pathogens producing more severe and prolonged recurrent mastitis [17,21].
Furthermore, by annotating the WMS reads against different reference databases such as the virulence factor database (VFDB) [24], ResFinder [25], KEGG and SEED modules [26,27], the study demonstrated that differences in mastitis-specific factors viz. VFGs, ARGs, and some metabolic functional potentials could strongly modulate microbiome dysbiosis and the pathophysiology of bovine mastitis (Figure 1). The study found the highest association of the VFGs in CM milk microbiomes followed by RCM, SCM and H milk microbes. The mastitis-associated pathogens harbored a wide range of VFGs, and bacteria having higher abundances also possessed increased number of associated VFGs in the corresponding metagenomes. As for example, mastitis causing microbiomes had sole association with several VFGs such as crucial nutrient (iron) uptake and/or regulation, twitching motility and/or chemotaxis, bacterial adhesion, invasion, or intracellular survival, biofilm formation, DNA uptake, adhesion to host cells and adherence, two-component system, intracellular multiplication factor, master regulator of biofilm initiation, quorum-sensing/antibiotics susceptibility altering, cellular adherence, heat shock stress-related, bacterial communication, and bacterial pathogenesis related. On the contrary, genes coding for bacterial chemotaxis, flagellar assembly, biofilm formation, alginate biosynthesis, pili and flagella expression/ adherence factors and outer membrane proteins had relative over expression in H milk microbiomes. The underlying mechanisms for microbial colonization in mammary tissue modulated by these VFGs are not well established. However, VFGs can enable microbiome to overcome host defense [2,28], immune-mediated colonization of the mammary tissues by suppressing the regrowth of resident commensal microbiota [13,29], and subsequent pathogenesis by sensing metabolites derived from the microbiota [29]. They may also be involved in the inclusion of opportunistic pathogens with the progression, and recurrence of the disease [2,29]. However, commensals can resist colonization of exogenous pathogens, and inhibit overgrowth of indigenous opportunists via several mechanisms, including metabolic competition [29].
The published study also stated the association of various homologues of ARGs belonged to different protein families among the microbiome of four metagenomes, and the composition and abundances of the ARGs remained significantly correlated with the abundances of the related bacteria. Of the detected ARGs, broad-spectrum beta-lactams, tetracyclines, aminoglycoside, sulfonamide, macrolides, fosfomycin and multidrug-resistant genes had several fold higher expressions in mastitis (CM, RCM, SCM) microbiomes compared to H milk microbiomes. Furthermore, the ARGs also varied greatly in different states of mastitis (CM, RCM, SCM) supporting the microbiome dysbiosis in the corresponding metagenomes, their genetic diversity, and selective pressure for the maintenance of ARGs [11]. The possible mechanisms for the detected ARGs include enzymatic inactivation, prevention of antibiotic-bindings to the targets, catalytic activity, folate pathway antagonist-attributed resistances and efflux pump and/or system conferring resistance to a wide range of antibiotics [11,30]. These ARGs can easily travel and spread throughout different dairy environments because of wind and runoff waters, climate change, human activities and contact with wild animals, in particular, migratory ones [2,30].
Discussion
This study also reported several important predicted metabolic functions that altered in their abundances between CM, RCM, SCM and healthy-milk microbiome as also reported previously in lactating cows [5,6,14-16], women [22,23] and mouse [18]. Our analysis revealed that similar metabolic features of microbiota between different states of mastitis are associated with their early colonization, and disease persistent and/or progression [12,31]. We found that metabolic functional pathways related to citrate metabolism (TCA cycle), citrate synthase, oxidative phosphorylation, methyl-accepting chemotaxis and two-component systems remained overexpressed in mastitis causing microbiomes. Conversely, the H milk microbiome had comparative overexpression in genes coding for pyruvate metabolism, cytochrome c oxidase subunits and protoheme IX farnesyltransferase. These altered metabolic pathways are associated with a diverse array of virulence mechanisms for mammary gland pathogenesis [13]. Bacterial biofilm-formation is a strain-specific or genetically-linked trait with selective advantage in pathogenesis of mastitis, and harmful to mammary tissues since they can promote the phagocyte release, proliferation of reactive oxygen and nitrogen species, and transfer of antibiotic resistance [2,5,6]. Moreover, mastitis causing microbiomes had overexpression of genes encoding biofilms adhesins [13,32], glutathione non-redox reactions [33], regulation of oxidative stress [34], and transposable elements [35] which play important role in many chronic-recurrent bacterial infections like bovine RCM.
Conclusion
This study conveyed that microbial changes in different types of mastitis resulted into depletion of beneficial microbes, and enrichment of opportunistic pathogens. The results of our published study suggest that co-occurrence of VFGs and ARGs might be taking place amid mastitis and healthy milk microbiomes. Several imputed functional pathways differed between mastitis and healthy-controls, possibly reflecting metabolic changes associated with mastitis pathogenesis. These findings will therefore reach a point to develop microbiomebased diagnostics and therapeutics for this economically burdened disease. However, future studies should delve with larger sample size with the inclusion of gut/rumen microbiome sampling in addition to the milk samples for direct testing of microbial transfer across this axis are needed to confirm the shift of microbiome and associated functions.
Acknowledgements
The Bangladesh Bureau of Educational Information and Statistics (BANBEIS), Ministry of Education, Government of Bangladesh (GoB) (Grant No. LS2017313) supported this work. The first author of this manuscript, MNH received his PhD Fellowship from the Bangabandhu Science and Technology Fellowship Trust, Ministry of Science and Technology, GoB. The authors thank Professor Keith Dr. A. Crandall, Founding Director, the Computational Biology Institute, Milken Institute School of Public Health, The George Washington University, USA for his support in sequencing. We also acknowledge the cooperation of the dairy farmers for allowing us to conduct the study in their farms.
REFERENCES
- Thompson-Crispi K, Atalla H, Miglior F, Mallard BA. Bovine mastitis: Frontiers in immunogenetics. Front Immunol. 2014; 5:493
- Hoque MN, Istiaq A, Rahman MS, Islam MR, Anwar A, Siddiki AZ, et.al. Microbiome dynamics and genomic determinants of bovine mastitis. Genomics. 2020; 112:5188-5203
- Jiménez E, de Andres J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, et al. Metagenomic analysis of milk of healthy and
- Jiménez E, de Andres J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, et al. Metagenomic analysis of milk of healthy and
- Sakwinska O. Bosco N. Host microbe interactions in the lactating mammary gland. Front Microbiol. 2019; 10:1863.
- Hoque MN, Clement RA, Sultana M, Crandall KA, Siddiki AZ, Hossain AM, et al. Metagenomic deep sequencing reveals association of microbiome signature with functional biases in bovine mastitis. Sci Rep. 2019; 9:13536.
- Hoque MN, Clement RA, Sultana M, Crandall KA, Siddiki AZ, Hossain AM, et al. Insights into the Resistome of Bovine Clinical Mastitis Microbiome, a Key Factor in Disease Complication. Front Microbial. 2020; 11:860.
- Oikonomou G, Addis FM, Chassard C, Macias MEF, Grant I, Delbes C, et al. Milk microbiota: what are we exactly talking about? Front Microbiol. 2020; 11:60.
- Bhatt VD, VB, PG, SJ, DN, DS, et al. Milk microbiome signatures of subclinical mastitis‐affected cattle analysed by shotgun sequencing. J Appl Microbiol. 2012; 112(4): 639–650.
- Hoque MN, Das ZC, Rahman A, Haider MG, Islam MA. Molecular characterization of Staphylococcus aureus strains in bovine mastitis milk in Bangladesh. Int. J Vet Sci Med. 2018; 6: 53–60.
- Hoque MN, Das ZC, Talukder AK, Alam MS, Abu Nasar Md, Rahman A,et al. Different screening tests and milk somatic cell count for the prevalence of subclinical bovine mastitis in Bangladesh. Trop Anim Health Prod. 2015; 47(1): 79-86.
- Escudeiro P, Pothier J, Dionisio F, Nogueira T. Antibiotic Resistance Gene Diversity and Virulence Gene Diversity Are Correlated in Human Gut and Environmental Microbiomes. mSphere. 2019; 4(3), 135-190.
- Derakhshani H, Fehr KB , Sepehri S, Francoz D , Buck JD , Barkema HW, Plaizier JC, et al. Invited review: microbiota of the bovine udder: contributing factors and potential implications for udder health and mastitis susceptibility. J Dairy Sci. 2018; 101(12):10605-10625.
- Gomes F, Maria JS, Mariana H. Bovine mastitis disease/pathogenicity: Evidence of the potential role of microbial biofilms. Pathogens Dis. 2016; 74(3).
- Cremonesi P, Pothier J, Dionisio F, Nogueira T, Sepehri S, Francoz D. et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS One. 2018; 13(10).
- Falentin H, Rault L, Nicolas A, Bouchard DS, Lassalas J, Lamberton P,et al. Bovine teat microbiome analysis revealed reduced alpha diversity and significant changes in taxonomic profiles in quarters with a history of mastitis. Front Microbiol. 2016; 7:480.
- Klaas IC, Zadoks RN. An update on environmental mastitis:
- Challenging perceptions. Transbound. Emerging Dis. 2018; 65:166 –185
- Ma C. Cow-to-mouse fecal transplantations suggest intestinal microbiome as one cause of mastitis. Microbiome .2018; 6(1):200
- Nayfach S. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019; 568:505–510.
- Maga EA, Weimer BC, Murray JD. Dissecting the role of milk components on gut microbiota composition. Gut Microbes. 2013; 4(2):136–139.
- Lurie-Weinberger, Gophna MN. Archaea in and on the human body: Health implications and future directions. PLoS Pathog. 2015; 11(6).
- Ruiz L, García-Carral C, Rodriguez JM. Unfolding the human milk microbiome landscape in the omics era. Front. Microbiol. 2019; 10.
- Patel S H,Sato Y, Furumichi M, Morishima K, Tanabe M,Gophna MN, et al. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep. 2017; 7(1):7804.
- Liu B, Zheng D, Jin Q, Chen L. Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019; 47: 687-692
- Doster E, García-Carral C, Rodriguez JM, Weimer BC, Murray JD, Thomason CA, et al. ResFinder 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2019; 48:561-569.
- Kanehisa M, Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M, et al. New approach for understanding genome variations in KEGG. Nucleic Acids Res.2019; 47:590–595.
- Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgunmetagenomes. Cold Spring Harb Protoc. 2010; 1:53-68
- Thomason CA, Mullen N, Belden LK, May M, Hawley DM, et al. Resident Microbiome Disruption with Antibiotics Enhances Virulence of a Colonizing Pathogen Sci Rep. 2017; 7.
- Addis MF, Tanca A, Uzzau S, Oikonomou G, Bicalho RC, Moroni P. The bovine milk microbiota: insights and perspectives from -omics studies. Mol Biosyst. 2016; 12:2359–2372.
- Van AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. Acquired antibiotic resistance genes: an overview. Front Microbiol. 2011; 2:203.
- Jamali H, Glass EM, Wilkening J, Wilke A, Antonopoulos D. Meyer F, Kanehisa M, et al. Invited review: Incidence, risk factors, and effects of clinical mastitis recurrence in dairy cows. J Dairy Sci. 2018; 101(6), 4729-4746.
- Fleitas MO,Cardoso MH, Meira S, Octavio R, Franco L, Bicalho RC, et al. Recent Advances in Anti-virulence Therapeutic Strategies with a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition. Front Cellular Infect Microbiol. 2019; 9:74.
- Kelly RA, Leedale J ,Calleja D, Steven J, Harrell A, Chadwick AE, Webb S, et al. Modelling changes in glutathione homeostasis as a function of quinone redox metabolism. Sci Rep. 2019; 9:6333.
Citation: Hoque MN, Sultana M, Hossain AM (2021) Dynamic Changes in Microbiome Composition and Genomic Functional Potentials in Bovine Mastitis. J Data Mining Genomics Proteomics. 12:232.
Copyright: © 2021 Hoque NM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

