Indexed In
- Open J Gate
- Genamics JournalSeek
- CiteFactor
- Cosmos IF
- Scimago
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- Directory of Abstract Indexing for Journals
- OCLC- WorldCat
- Proquest Summons
- Scholarsteer
- ROAD
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 15, Issue 5
Diagnosis of Colon Cancer Using LncRNA ROR Gene Biomarker
Maria Beihaghi1*, Reza Sahebi2, Mohammad Reza Beihaghi3, Raheleh Khosravi Nessiani4 and Majedeh Ramian52Department of Modern Sciences and Technologies, Mashhad University of Medical Sciences, Mashhad, Iran
3Department of Sociology and Politics, Sheffield Hallam university, Sheffild, England
4Department of Chemical Engineering, Sahand University of Technology, Tabriz, Iran
5Department of Biotechnology, Ferdowsi University of Mashhad, Mashhad, Iran
Received: 23-Dec-2022, Manuscript No. BLM-23-19451; Editor assigned: 26-Dec-2022, Pre QC No. BLM-23-19451 (PQ); Reviewed: 09-Jan-2023, QC No. BLM-23-19451; Revised: 08-Mar-2023, Manuscript No. BLM-23-19451 (R); Published: 15-Mar-2023, DOI: 10.35248/0974-8369.23.15.556
Abstract
Colorectal Cancer (CRC), which is one of the most commonly diagnosed cancers worldwide, threatens human health. Since the pathological mechanism of CRC is not entirely understood, there are many challenges in preventing, diagnosing, and treating this disease. Long non-coding RNAs (LncRNAs) have recently gained attention due to their potential role in various stages of CRC formation, invasion, and progression, including regulation of molecular signaling pathways, apoptosis, autophagy, angiogenesis, tumor metabolism, immune responses, cell cycle, and Epithelial Mesenchymal Transition (EMT). The role of long non-coding RNA Regulator of Reprogramming (ROR) in cellular reprogramming has been studied in this research. After extracting and preparing RNA from 100 cancerous and 100 non-cancerous tissues, the expression changes of Linc-ROR and all samples were analyzed with the Real time PCR technique. The statistical test results showed that Linc-ROR has a significant expression increase in tumor samples compared to non-tumor samples. According to the obtained results, the expression of the Linc- ROR gene is expected to be related to tumorigenesis and is very important in inhibiting apoptosis during cell stress.
Keywords
Non-coding RNAs; Colon cancer; Real time PCR; Linc-RNA; ROR
Introduction
Colorectal Cancer (CRC) is one of the most common cancers diagnosed worldwide. In the United States, CRC is the third leading cause of cancer related death in men and women. There will be over 10,000 new cases in 2020, and the proportion of young patients is increasing [1]. In addition, the incidence of CRC in developing countries is approximately 4 times higher than in developing countries [2]. In Iran, colon cancer is among the five most common cancers, with a high prevalence in the north. According to the ministry of health, Iranian men had 9.27, 9.64, and 9.90 deaths per 100,000 people in 2012, 2013, and 2015, while Iranian women had 9.12, 9.47, and 9.13 deaths per 5,000 people in each year [3].
Besides a family history of cancer and inherited conditions, factors such as obesity, physical inactivity, alcohol intake, processed foods, and smoking can increase the risk of colon cancer. Several methods are used to diagnose this disease, including guaiac fecal occult blood testing, sigmoidoscopy, colonoscopy, and fecal immunochemical tests [4]. The lack of noninvasive and low-cost prognostic and diagnostic tests for CRC has increased interest in identifying novel, potentially effective biomarkers [5]. Since the pathological mechanism of CRC is not yet fully understood, further studies are necessary to identify and develop new effective biomarkers and targets for its diagnosis and treatment.
Long non-coding RNAs (LncRNAs) are transcripts with a length of more than 200 nucleotides without the ability to encode proteins, which play different roles in regulating gene expression at the transcription and translation levels, as well as epigenetic changes [6]. Various diseases, including cancer, are associated with dysfunctions in LncRNAs expression or function [7]. It has been found that these long non-coding transcripts, which have unique profiles in multiple cancers, are involved in the inhibition of programmed death, proliferation, migration, and invasion of cancer cells to other tissues [8]. Lncrnas, like other cancer related nucleic acid fragments released from cells, are introduced into the bloodstream of cancer patients and provide the possibility of non-invasive evaluation of gene expression. For this reason, these non-coding transcripts can be used as biomarkers for the prognosis and diagnosis of cancer. Recent studies have shown that the expression of LncRNAs in some human tumors, including colon cancer (CRC), is significantly increased or inhibited [9].
LincRNA-ROR was first identified in 2010 when studying the reprogramming of fibroblast cells into induced pluripotent cells. If LncRNA-ROR is inhibited, the reprogramming process in these cells is inhibited [10].
In 2014, Hu, et al. revealed that Linc-ROR plays a role in breast cancer metastasis and causes breast cancer cells' invasive and migratory state. Furthermore, Linc-ROR causes Epithelial to Mesenchymal Transformation (EMT) through miR-205 absorption and removes its inhibitory effect from ZEB1 and ZEB2 factors in breast cancer [11]. In other studies conducted by Zheng, et al. Linc-ROR acts as a repressor for p53 protein in stressful conditions and inhibits the p53 dependent apoptosis pathway. The mechanism of action of ROR is through posttranscriptional processes so that the 28 bp sequence in the fourth exon of Linc-ROR has a motif that binds to hnRNP-1, which is necessary for p53 inhibition. In other words, hnRNP-1 binds to mRNA transcribed from the p53 gene and prevents translation [12].
Takahashi, et al. have shown that the number of ROR-positive liver cancer cells is higher in hypoxic areas than in non-hypoxic areas. Their other research showed that Linc-ROR changes the expression level of HIFI-a factor, which is mediated by miR-145. The expression of this factor is known as a prognostic factor and increases in aggressive tumors [13].
Considering the high rate of incidence and mortality caused by colon cancer and the critical role of ROR in the formation and development of this cancer, and since there has been no research on the relationship between the expression levels of ROR and susceptibility to colon cancer, in this research, the study of the level The expression of Linc-RNA ROR was analyzed. LncRNAs can regulate CRC occurrence and development. Some LncRNAs tend to cancer cell proliferation, invasion, metastasis, and drug resistance; some suppress cancer cell proliferation and metastasis. In the following, we will discuss the relationship between this biomarker and colon cancer using the analyzes done.
Materials and Methods
Sample collection
In this research, to investigate the expression changes of the Linc-RNA ROR encoding gene, 200 samples of tumor and nontumor tissues were prepared as purified RNA from Bu Ali research institute of Mashhad. None of the selected patients had been subjected to various treatments, such as chemotherapy or radiation therapy. According to the written permission form, all the people whose tissue samples were used in this research declared their consent to conduct this study.
RNA extraction
RNA was extracted from the entire cell using trizol (Invitrogen, USA). 1% agarose gel was used to determine the quality of RNA extraction; then, the samples were treated with 2 microliters of DNaseI (Japan, Bio TaKaRa) enzyme for 30 minutes at 37℃ to remove DNA contamination. RNA quantity and quality were determined by the optical density method using a nanodrop (2000 nanodrop scientific thermo) device.
Primer design and cDNA synthesis
The expression level of the LncRNA-ROR encoding gene was investigated by the real time PCR. To check the expression of the target gene, specific primers for each cDNA sequence were designed using per primer V 4.1.21 software, oligo V 7.5b, and generunner V 3.5. The primer used for Linc-ROR, Linc-ROR variants, and the internal primers (GAPDH) is shown in Table 1.
| Gene | Primer | Sequence |
|---|---|---|
| Linc-ROR | F | ACAAGGAGGAAAGGGCTGAC |
| R | TTCTGGAAGCTAAGTGCACATG | |
| GAPDH | F | ATGGGGAAGGTGAAGGTCG |
| R | GGGGTCATTGATGGCAACAATA |
Table 1: Primer sequence used for linc-ROR and internal primer (GAPDH) as a reference gene.
With a total volume of 40 microliters, a real time PCR reaction mixture was prepared using 10 microliters of cybergreen mix (Takara Bio, Japan); 1 microliter of cDNA (concentrated at 100 ng/microliter), 10 picomoles of reverse primer mixture, and 19 microliters of nuclease free water. Polymerase chain reaction performed in 40 cycles, according to the timetemperature schedule in Table 2, using (applied biosystem step one) device.
Statistical analysis
Real time-PCR data was analyzed using LinReg PCR software. The ΔΔCT method was used to investigate the relative changes in gene expression. Finally, graph pad prison V 5.0.0288 software was used for data analysis and ROC construction to describe the specificity and sensitivity of the diagnosis.
Results
Determination of quantity and quality of extracted RNA
Electrophoresis, agar gel, and spectrophotometry were used to ensure the integrity and quality of the extracted RNA. The creation of two bands of 28 S and 18 S on agarose gel showed that RNA was not degraded. In addition, the smear formed between two bands indicates the presence of mRNA. In addition, the optical absorption ratio of 260/280 is about 2-1.9 in the spectrophotometry analysis, showing that the RNA has a high purity percentage (Figure 1).
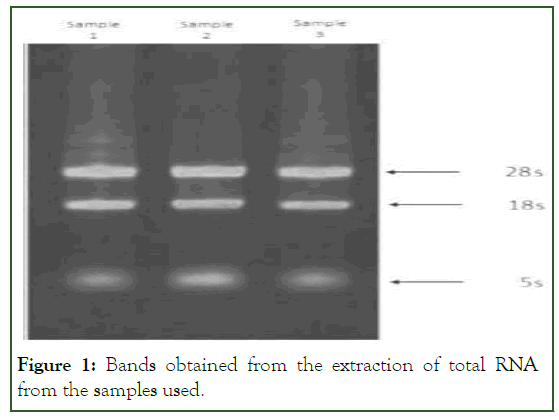
Figure 1: Bands obtained from the extraction of total RNA from the samples used.
Semi-quantitative analysis of gene expression by RTPCR
To check the correctness and accuracy of the primers in correctly identifying the amplified region, the absence of contamination with DNA and dimer was checked using PCR. According to the gel electrophoresis and obtaining a single band, no DNA contamination and specificity were observed. Then, based on the GAPDH standard band, the balance of bands 28 and 18 of ribosomal RNA was carried out after quantifying the RNA bands, cDNA using specific primers of the Linc-ROR gene (Table 2), entered the RT-PCR reaction and A fragment of 124 expected game pairs was observed (Figure 2).
| Phase | Temperature (℃) | Time | Number of each cycle |
|---|---|---|---|
| Internal denaturation | 95 | 5 min | 1 |
| Denaturation | 95 | 30 s | 30-38 |
| Connection | 63 | 30 s | |
| Expansion | 72 | 30 s | |
| Final expansion | 72 | 15 min | 1 |
Table 2: PCR reaction timetable.
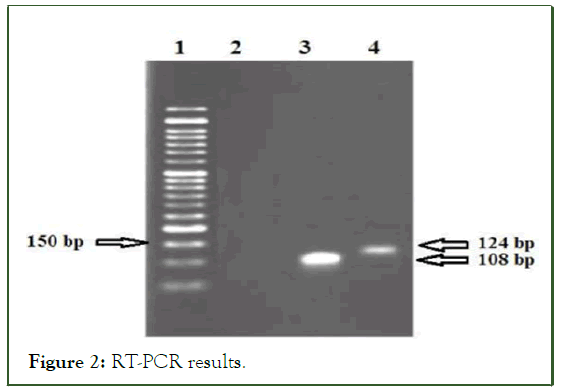
Figure 2: RT-PCR results.
Band quantification analysis was performed using lab works and total lab software. Each sample's quantity of reproduced band was determined relatively compared to the band of 400 pairs of markers (commercial pars twin).
Determining primer efficiency
Linreg PCR software was used to check the efficiency of the primer. After examining the efficiency of the primer, the final average showed in Table 3.
| Gene | Linc-ROR |
|---|---|
| Efficiency mean | 1.892 |
Table 3: Determining the efficiency of linc-ROR primer.
Results of real-time PCR reaction
The real-time PCR reaction was performed, and the results were recorded. The relative level of Linc-ROR gene expression was obtained by analyzing the CT values using REST and excel software. A kolmogorov-smirnov test was used to determine normality, and then a T-test was done based on that. AT student analysis revealed that the Linc-ROR gene has a significant expression level (p<0.05). Then the obtained data was used for the final analysis using graph pad software. According to the results of the Linc-ROR gene expression, it has a significant level in tumor and non-tumor samples (p<0.05) (Figure 3). Based on the degree of malignancy, it was divided into two groups, low and high (grades II and III) (Figure 4).
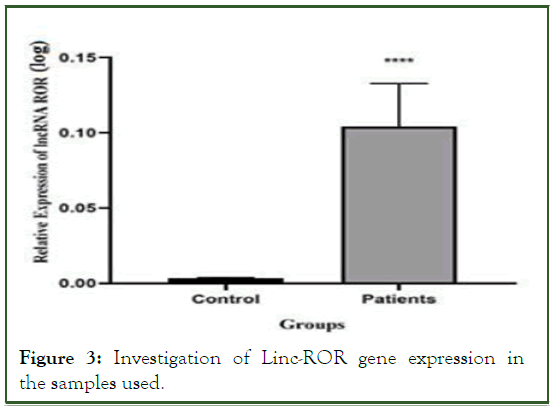
Figure 3: Investigation of Linc-ROR gene expression in the samples used.
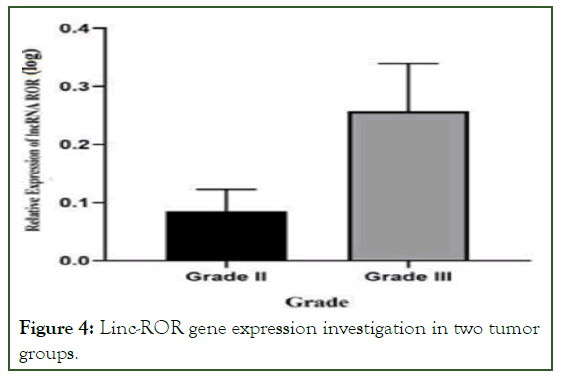
Figure 4: Linc-ROR gene expression investigation in two tumor groups.
Results of the ROC curve analysis
The Linc-ROR gene expression results were analyzed using graph pad prism version 8 software. The efficiency of gene expression for becoming a biomarker was checked. The results of the ROC curve show that the expression of the desired gene has a solid potential for becoming a biomarker. The Area Under the Curve (AUC) shows specificity above 80%.
Melting temperature curve of linc-ROR and GAPDH
The curve of the maximum melting temperature was obtained for each of the two pairs of Linc-ROR and GAPDH specific primers, which showed that the primers did not interact with each other and that the reaction had a suitable efficiency for determining gene expression quantification. In addition, according to the curve, it can be concluded that the primers acted specifically and single samples of cDNA have been amplified (Figure 5).
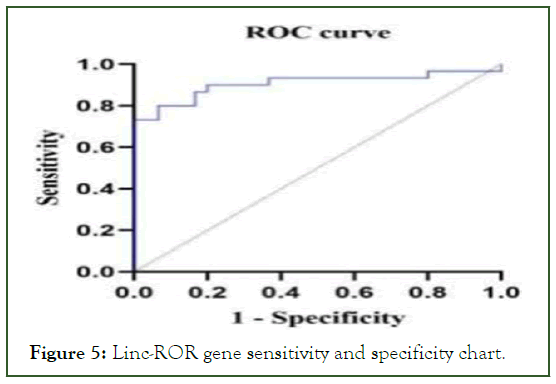
Figure 5: Linc-ROR gene sensitivity and specificity chart.
Discussion
In this study, the expression level of the LncRNA-ROR gene in the cells of cancer tissue and the adjacent normal tissue of colon cancer was evaluated and compared. The results showed a significant increase in LncRNA-ROR in each sample studied in tumor tissue compared to normal tissue.
Long Non-Coding RNAs (LncRNAs) regulate their expression by targeting the non-coding sequences of the 3 sides of their target genes [14]. Previous studies have shown that the activity of about 18% of non-coding genes of LncRNAs is related to various cancers. In comparison, only 9% of human protein coding genes are related to multiple cancers [15]. The evidence shows that the change in the expression of LncRNAs is associated with tumor formation [16].
In recent years, it has been determined that the abnormal expression of some LncRNAs plays an important role in the formation and progression of colon cancer. For example, it has been found that increasing the expression of LncRNAs CCAT1-1, MALAT1, PVT, and HOTAIR in colon cancer cells can play an essential role in tumor formation and metastasis to other tissues [17,18].
In 2013, studies on the molecular mechanism of Linc-RNA showed that Linc-ROR removes the inhibitory effect of miR-145 on pluripotency maintenance factors such as Sox2, Nanog and Oct4 by acting like a sponge. Also, inhibiting Linc-ROR leads to the differentiation of embryonic stem cells into mesoderm and ectoderm layers [19]. Several signaling pathways are mediated by Lincrnaror in tumor growth and progression. By regulating the TGF-b pathway, LncRNAs-ROR promotes the proliferation, migration, and invasion of breast cancer [20]. In one of the metaanalysis studies conducted by Quan, et al. the importance of the expression of several LncRNA and their relationship with bladder cancer has been investigated. The results show a significant relationship between lymph node metastasis and the expression of several LncRNA, including SPRY4-IT1, UBC1, and ROR. These findings suggest that LncRNAs can be used as biomarkers of lymph node metastasis. Furthermore, in a meta data-analysis obtained from 903 patients with cancer in 11 studies, it was shown that there is a positive correlation between ROR gene expression and tumor invasion depth, Lymph Node Metastasis (LNM) TNM stage, Overall Survival (OS), and Disease Free Survival (DFS). Based on the Gao, et al. article, Lnc-ROR can be introduced as a diagnostic biomarker for pancreatic cancer. Accordingly, silencing this gene can suppress proliferation, invasion, and tumorigenicity, leading to the decline of malignancy in Pancreatic Cancer Stem Cells (PCSCs).
As a prognostic tool, a biomarker should be highly sensitive and show a significant relationship with the pathological aspects of the disease. Triple negative breast cancer has had difficulty being identified due to the absence of Estrogen Receptor (ER), Progesterone Receptor (PR), and Human Epidermal growth factor Receptor 2 (HER2) receptors. Despite the aggressiveness and rapid progression of TNBC, it is crucial to use the most reliable diagnostic methods. Eades, et al. determined that miR-145 affects TNBC cell invasion by trans well invasion assays. This study found that mature miR-145 is down regulated by the LincrRNA-ROR, acting as a sponge competitive endogenous RNA (ceRNA). Another study investigated expression levels of LncRNA-ROR in 36 patients with Renal Cell Carcinoma (RCC) with qRT-PCR techniques. Compared to adjacent non-tumor tissues, RCC tissues showed a higher expression of LncRNA ROR. Moreover, at clinical stages III and IV, the expression of LncRNA ROR was higher than at clinical stages I and II. According to another research study on breast cancer, Lnc-ROR functions as an oncogene. It also provides evidence that LncRNAs ROR decoys transmethylase MLL1 to promote H3K4 methylation and regulate TIMP3, thereby increasing breast cancer progression.
Based on our findings in this study, expression of the LNC-ROR gene in tumor tissue samples was remarkedly higher than in normal tissues. Additionally, we demonstrated that this gene is expressed at a greater level in clinical stage III than in stage II. It is consistent with the findings of studies discussed earlier in this article that a higher malignancy of the cancer tissue is associated with a higher expression of ROR.
Conclusion
Research efforts are always directed toward finding the most effective treatment for diseases, including colon cancer. Identifying the proper treatment requires a correct diagnosis. Selecting biomarkers for disease diagnosis has become one of the most important strategies in recent years. Investigating the expression levels of LncRNA ROR in tumor and normal samples of colon cancer patients using the real time-PCR method showed a significant difference between the expression levels of this LncRNA in the two sample groups. Results show that tumor cells have a higher level of long non-coding RNA expression than normal cells. Moreover, Linc-ROR gene expression is higher in grade III than in grade II. Therefore, it may be possible to measure the amounts of this LncRNA in the peripheral blood of people who are prone to or suspected of colon cancer as a non-invasive method for early diagnosis of the disease. Further studies are necessary to clarify Linc-ROR's role as a biomarker in colon cancer.
References
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145-164.
[Crossref] [Google Scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;1(3):209-249.
[Crossref] [Google Scholar] [PubMed]
- Haggar FA, Boushey RP. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clinics Colon Rectal Surgery. 2009;22(4):191.
[Crossref] [Google Scholar] [PubMed]
- Johnson CM, Wei C, Ensor JE, Smolenski DJ, Amos CI, Levin B, et al. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control. 2013;24:1207-1222.
[Crossref] [Google Scholar] [PubMed]
- Chaleshi V, Irani S, Alebouyeh M, Mirfakhraie R, Aghdaei HA. Association of lncRNA-p53 regulatory network (lincRNA-p21, lincRNA-ROR, and MALAT1) and p53 with the clinicopathological features of colorectal primary lesions and tumors. Oncol Lett. 2020;19(6):3937-3949.
[Crossref] [Google Scholar] [PubMed]
- Siddiqui H, Al-Ghafari A, Choudhry H, Al Doghaither H. Roles of long non-coding RNAs in colorectal cancer tumorigenesis: A review. Mol Clin Oncol. 2019;11(2):167-172.
[Crossref] [Google Scholar] [PubMed]
- Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9(7):1354-1366.
[Google Scholar] [PubMed]
- Galamb O, Bartak BK, Kalmar A, Nagy ZB, Szigeti KA, Tulassay Z, et al. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol. 2019;25(34):5026-5048.
[Crossref] [Google Scholar] [PubMed]
- Poursheikhani A, Abbaszadegan MR, Kerachian MA. Long non-coding RNA AC087388.1 as a novel biomarker in colorectal cancer. BMC Cancer 2022;22:196.
[Crossref] [Google Scholar] [PubMed]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-ROR modulates reprogramming of human induced pluripotent stem cell. Nat Genet. 2010;42(12):1113-1117.
[Crossref] [Google Scholar] [PubMed]
- Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. LincRNA-ROR induces epithelial to mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5(6):e1287.
[Crossref] [Google Scholar] [PubMed]
- Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23(3):340-50.
[Crossref] [Google Scholar] [PubMed]
- Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127(7):1585-1594.
[Crossref] [Google Scholar] [PubMed]
- Khachane AN, Harrison PM. Mining mammalian transcript data for functional long noncoding RNAs. PloS One. 2010;5(4):e10316.
[Crossref] [Google Scholar] [PubMed]
- Oliva J, Bardag-Gorce F, French BA, Li J, et al. The regulation of non-coding RNA expression in the liver of mice fed DDC. Exp Mol Pathol. 2009;87(1):12-19.
[Crossref] [Google Scholar] [PubMed]
- Li DX, Fei XR, Dong YF, Cheng CD, Yang Y, Deng XF, et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8(50):88163-88178.
[Crossref] [Google Scholar] [PubMed]
- Nissan A, Stojadinovic A, Mitrani-Rosenbaum S, Halle D, Grinbaum R, Roistacher M, et al. Colon cancer associated transcript-1: A novel RNA expressed in malignant and pre-malignant human tissues. Int J Cancer. 2012;130(7):1598-1606.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69-80.
[Crossref] [Google Scholar] [PubMed]
- Hou L, Tu J, Cheng F, Yang H, Yu F, Wang M, et al. Long noncoding RNA ROR promotes breast cancer by regulating the TGF-beta pathway. Cancer Cell Int. 2018;18:142.
[Crossref] [Google Scholar] [PubMed]
- Quan J, Pan X, Zhao L, Li Z, Dai K, Yan F, et al. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: A systematic review and meta-analysis. Onco Targets Ther. 2018;11:6415-6424.
[Crossref] [Google Scholar][PubMed]
Citation: Beihaghi M, Sahebi R, Beihaghi MR, Nessiani RK, Ramian M (2023) Diagnosis of Colon Cancer Using LncRNA ROR Gene Biomarker. Bio Med. 15:556.
Copyright: © 2023 Beihaghi M, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


