Indexed In
- Academic Journals Database
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 15, Issue 2
Comparative Studies and Evolution of Mammalian and Bird CA1, CA2, CA3 and CA13 Genes and Proteins
Roger S Holmes1,2*2Griffith Research Institute for Drug Design, Griffith University, Nathan, Australia
Received: 08-Apr-2024, Manuscript No. JDMGP-24-25434 ; Editor assigned: 10-Apr-2024, Pre QC No. JDMGP-24-25434 (PQ); Reviewed: 24-Apr-2024, QC No. JDMGP-24-25434 ; Revised: 01-May-2024, Manuscript No. JDMGP-24-25434 (R); Published: 08-May-2024, DOI: 10.4172/2153-0602.24.15.338
Abstract
Mammalian carbonic anhydrases (E.C.4.2.1.2; CA , Ca, Cah or CAH) genes encode enzymes that catalyse the reversible hydration of carbon dioxide and contribute significantly to many other biological phenomena. CA genes and enzymes from several mammalian species which have been assigned to at least 15 gene families, including CA1-3 and CA13, which are closely localized within a gene complex on human chromosome 8. This paper reports the amino acid sequences, gene locations, tissue expression patterns and exon structures for mammalian CA1, CA2, CA3 and CA13 genes and proteins, including primates, other eutherian mammals and a marsupial mammal. The phylogenetic and evolutionary relationships of these genes and enzymes are described with a hypothesis for gene duplication events for ancestral mammalian CA1, CA2, CA3 and CA13 genes, generating 4 families of these genes, which are closely localized on mammalian genomes and are differentially expressed in tissues of the body.
Keywords
Human; Mouse; Mammals; Bird; Carbonic anhydrases; Gene complex; Enzymes; Evolution
Introduction
At least fifteen families of mammalian Carbonic Anhydrase genes (CAfor humans and primates; Car for mouse and rat) and enzymes (CA; CAR; or CAH; E.C.4.2.1.2; also called carbonate dehydratases) have been recognized by the respective human (genenames.org) and mouse (informatics.jax.org) gene nomenclature authorities. These include: CA1, encoding the major erythrocyte enzyme [1,2]; CA2, the major intestinal enzyme [3,4]; CA3, the major enzyme in red skeletal muscle [5,6]; and CA13, with a widespread distribution pattern in human tissues [7,8]. These enzymes catalyze the reversible hydration of carbonic dioxide and contribute significantly to many other biological phenomena, including the formation of body fluids (gastric acid, aqueous humor, cerebrospinal fluid and saliva), respiration, bone resorption, calcification, intracellular pH regulation and chloride-bicarbonate exchange activity [9,10].
Structures for several human and animal CA1-3 and CA13 zinc metalloenzymes proteins have been reported, including human CA1 (Pdb:1AZM) [1,11]; CA2 (Pdb:12CA) [3,4]; CA3 (Pdb:1Z93) [5]; and CA13 (Pdb:3CZV) [8]. In addition, variants of CA have also been associated with human diseases, including atherosclerosis, cancer, obesity, epilepsy, edema and glaucoma and are the subject of extensive drug research [9,10,12,13]. Genetic analyses of CA1- 3 in humans and mice have reported that these genes are closely localized on chromosomes 8 and 3, respectively [14-16]. Subsequent studies have incorporated CA13 into the CA1-3 and CA13 human and mouse CA gene complex [17]. This paper reports the predicted amino acid sequences, gene locations, tissue expressions and exon structures for mammalian CA1, CA2, CA3 and CA13 genes and proteins, including primates, other eutherian mammals and a marsupial mammal. The phylogenetic and evolutionary relationships of these genes and enzymes are described with a hypothesis for gene duplication events for ancestral mammalian CA1, CA2, CA3 and CA13 genes, generating 4 families of these genes, which are closely localized on mammalian genomes and are differentially expressed in tissues of the body.
Materials and Methods
CA1, CA2, CA3 and CA13 gene and protein identification
BLAST (Basic Local Alignment Search Tool) studies were undertaken using web tools from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [18]. BLAST analyses used the reported human CA1, CA2, CA3 and CA13 amino acid sequences [1,3,5,8]. Non-redundant mammalian protein sequence databases were analyzed using the blastp algorithm [18]. BLAT analyses were subsequently undertaken for each of the predicted CA1, CA2, CA3 and CA13 amino acid sequences using the UC Santa Cruz web browser [19] [http://genome.ucsc.edu/cgi-bin/hgBlat] to obtain the predicted locations for each of the mammalian and other vertebrate CA genes, including exon boundary locations and gene sizes (Table 1). Genomic sequences studied included: Human (Homo sapiens) [20]; Rhesus monkey (Macaca mulatta) [21]; African green monkey (Chlorocebus aethiops sabeus) [22]; Mouse (Mus musculus) [23]; Cow (Bos taurus) [24]; Opossum (Monodelphis domestica) [25]; and brown kiwi (Apteryx mantelli) [26]. Structures for the major isoforms of human CA1, CA2, CA3 and CA13 were obtained using the AceView website to examine predicted gene and protein structures to interrogate this database of human mRNA sequences (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) [27].
| Species | CA | Gene location | Transcript ID | Exon (strand) | UNIPROT ID | Amino acids |
|---|---|---|---|---|---|---|
| Human | CA13 | 8:85,245,829-85,281,346 | BC052602 | 7 (+) | Q8N1Q1 | 262 |
| CA1 | 8:85,328,563-85,338,450 | BC827890 | 7 (-) | P00915 | 261 | |
| CA3 | 8:85,438,910-85,448,150 | BC004897 | 7 (+) | P07451 | 260 | |
| CA2 | 8:85,465,271-85,480,786 | M77180 | 7 (+) | P00918 | 260 | |
| Rhesus monkey | CA13 | 8:85,792,235-85,826,005 | XP_001095487 | 7 (+) | A0A1D5QB60 | 262 |
| CA1 | 8:85,874,315-85,884,351 | XP_015001152 | 7 (-) | P00916 | 261 | |
| CA3 | 8:85,982,152-85,990,512 | XP_015001153 | 7 (+) | F6TQ33 | 260 | |
| CA2 | 8:86,007,619-86,023,635 | NM_00195417 | 7 (+) | F6TQ14 | 260 | |
| Green monkey | CA13 | 8:80,617,898-80,651,689 | XP_007999191 | 7 (+) | A0A0D9RL50 | 262 |
| CA1 | 8:80,699,654-80,709,377 | XP_007999193 | 7 (-) | A0A0D9RL45 | 261 | |
| CA3 | 8:80,806,899-80,815,255 | XP_007999197 | 7 (+) | A0A0D9RL40 | 260 | |
| CA2 | 8:80,831,138-80,848,058 | XP_007999199 | 7 (+) | A0A0D9RL35 | 260 | |
| Mouse | Ca13 | 3:14,645,036-14,661,571 | AK162621 | 7 (+) | Q9D6N1 | 262 |
| Ca1 | 3:14,766,539-14,778,384 | NM_009799 | 7 (-) | P13634 | 261 | |
| Ca3 | 3:14,864,249-14,871,658 | BC011129 | 7 (+) | P16015 | 260 | |
| Ca2 | 3:14,887,833-14,900,087 | NM_009801.4 | 7 (+) | P00920 | 260 | |
| Cow | CA13 | 14:77,334,460-77,371,694 | BC103269 | 7 (-) | A0A3Q1NEZ9 | 262 |
| CA1 | 14:77,194,103-77,204,445 | BC116126 | 7 (+) | Q1LZA1 | 261 | |
| CA3 | 14:77,029,841-77,038,943 | BC102666 | 7 (-) | Q3SZX4 | 260 | |
| CA2 | 14:76,995,220-77,010,922 | BC103269 | 7 (-) | P00921 | 260 | |
| Opossum | CA13 | 3:145,719,578-145,783,031 | XP_001366749 | 7 (-) | A0A5F8GLC4 | 263 |
| CA1 | 3:145,599,056-145,608,788 | AJ417908 | 7 (+) | Q8HY33 | 262 | |
| CA3 | 3:145,478,524-145,497,885 | XP_001366645 | 7 (-) | F6U1Y6 | 260 | |
| CA2 | 3:145,417,879-145,439,262 | XP_001376657 | 7 (-) | na | 265 | |
| Kiwi | CA13 | *3,710,625-3,725,122 | XP_025931592 | 7 (-) | na | 258 |
| CA1 | *3,671,191-3,680,784 | XP_025931593 | 7 (+) | na | 259 | |
| CA3 | *3,580,584-3,600,882 | XP_013812316 | 7 (-) | na | 265 | |
| CA2 | *3,491,289-3,514,049 | XP_025931607 | 7 (-) | na | 260 | |
| *NW_014004943v1 |
Table 1: Mammalian and bird CA1, CA2, CA3 and CA13 genes and subunits. Transcript IDs, GenBank and UNIPROT IDs provide the sources for the gene and protein sequences; +ve and –ve refer to the transcription strand; brown kiwi (Apteryx rowi) CA genes were located within a gene complex on chromosomal segment NW_01400943v1 [26].
Predicted structures and properties of CA1, CA2, CA3 and CA13 subunits
Alignments of predicted CA1, CA2, CA3 and CA13 amino acid sequences and estimates of sequence identities were undertaken using a ClustalW method (http://www.ebi.ac.uk/Tools/msa/ clustalw2/) [28]. Secondary structures for human CA subunits were obtained from the reported tertiary structures for human CA1 [1]; CA2 [3]; CA3 [5]; and CA13 [8].
Human CA1, CA2, CA3 and CA13 gene expression and predicted gene regulation sites
The GTEx web browser (http://gtex.org) was used to examine the human tissue expression profiles for CA1, CA2, CA3 and CA13 genes [29]. The human genome browser (http://genome.ucsc.edu) was used to examine predicted CpG islands [30], and Transcription Factor Binding Sites (TFBS) (ORegAnno IDs: Open Regulatory Annotations) [31], for human CA1, CA2, CA3 and CA13 genes using the UC Santa Cruz Genome Browser [32].
Phylogenetic studies and sequence divergence
Mammalian and bird (brown kiwi) (Apteryx mantelli) CA1, CA2, CA3 and CA13 amino acid sequences were subjected to phylogenetic analysis using the http://www.phylogeny.fr/ portal to enable alignment (MUSCLE), curation (Gblocks), phylogeny (PhyML) and tree rendering (TreeDyn) to reconstruct phylogenetic relationships [33]. Mammalian and bird (brown kiwi) CA sequences were identified as members of the CA1, CA2, CA3 or CA13 groups of enzymes.
Results and Discussion
Alignments and biochemical features of CA1, CA2, CA3 and CA13 amino acid sequences
Amino acid sequence alignments for human CA1, CA2, CA3 and CA13 amino acid sequences are shown in Figure 1, together with the reported secondary structure and key amino acid residues for CA1 [1], CA2 [3]; CA3 [5]; and CA13 [8]. The human CA1, CA2, CA3 and CA13 sequences shown exhibited>50% identities, suggesting that these protein subunits are products of a single gene family, but with 78% or more sequence identities, comparing human, rhesus monkey and mouse CA sequences within the same family group (Table 2). Amino acid sequences for the eutherian mammalian CA proteins examined contained 261 (CA1), 260 (CA2 and CA3), and 262 (CA13) residues (Table 1) whereas the corresponding opossum (Monodelphis domestica) and brown kiwi (Apteryx rowi) CA sequences contained similar numbers of amino acids to those for eutherian mammals (Table 1).
| Human CA1 | Rhesus CA1 | Mouse CA1 | Human CA2 | Rhesus CA2 | Mouse CA2 | Human CA3 | Rhesus CA3 | Mouse CA3 | Human CA13 | Rhesus CA13 | Mouse CA13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human CA1 | 100 | 95 | 78 | 60 | 60 | 59 | 54 | 54 | 55 | 60 | 60 | 64 |
| Rhesus CA1 | 95 | 100 | 78 | 60 | 60 | 59 | 54 | 54 | 55 | 60 | 60 | 60 |
| Mouse CA1 | 78 | 78 | 100 | 59 | 59 | 58 | 55 | 55 | 56 | 61 | 62 | 64 |
| Human CA2 | 60 | 60 | 59 | 100 | 98 | 81 | 58 | 58 | 59 | 60 | 59 | 61 |
| Rhesus CA2 | 60 | 60 | 60 | 98 | 100 | 81 | 59 | 59 | 62 | 60 | 59 | 61 |
| Mouse CA2 | 54 | 54 | 58 | 81 | 81 | 100 | 56 | 56 | 57 | 60 | 59 | 57 |
| Human CA3 | 60 | 60 | 55 | 58 | 58 | 56 | 100 | 96 | 91 | 58 | 58 | 59 |
| Rhesus CA3 | 59 | 59 | 55 | 59 | 59 | 56 | 96 | 100 | 92 | 57 | 58 | 57 |
| Mouse CA3 | 60 | 60 | 64 | 59 | 58 | 57 | 91 | 92 | 100 | 59 | 59 | 60 |
Human CA13 |
60 | 60 | 61 | 60 | 60 | 57 | 58 | 58 | 57 | 100 | 96 | 91 |
| Rhesus CA13 | 60 | 60 | 62 | 60 | 59 | 59 | 58 | 58 | 59 | 96 | 100 | 92 |
| Mouse CA13 | 64 | 60 | 64 | 61 | 61 | 57 | 59 | 57 | 60 | 91 | 92 | 100 |
Note: Numbers in bold show higher sequence identities for the more closely related CA family members.
Table 2: Percentage identities for mammalian CA1, CA2, CA3 and CA13 amino acid sequences. Numbers show the percentage of amino acid sequence identities.
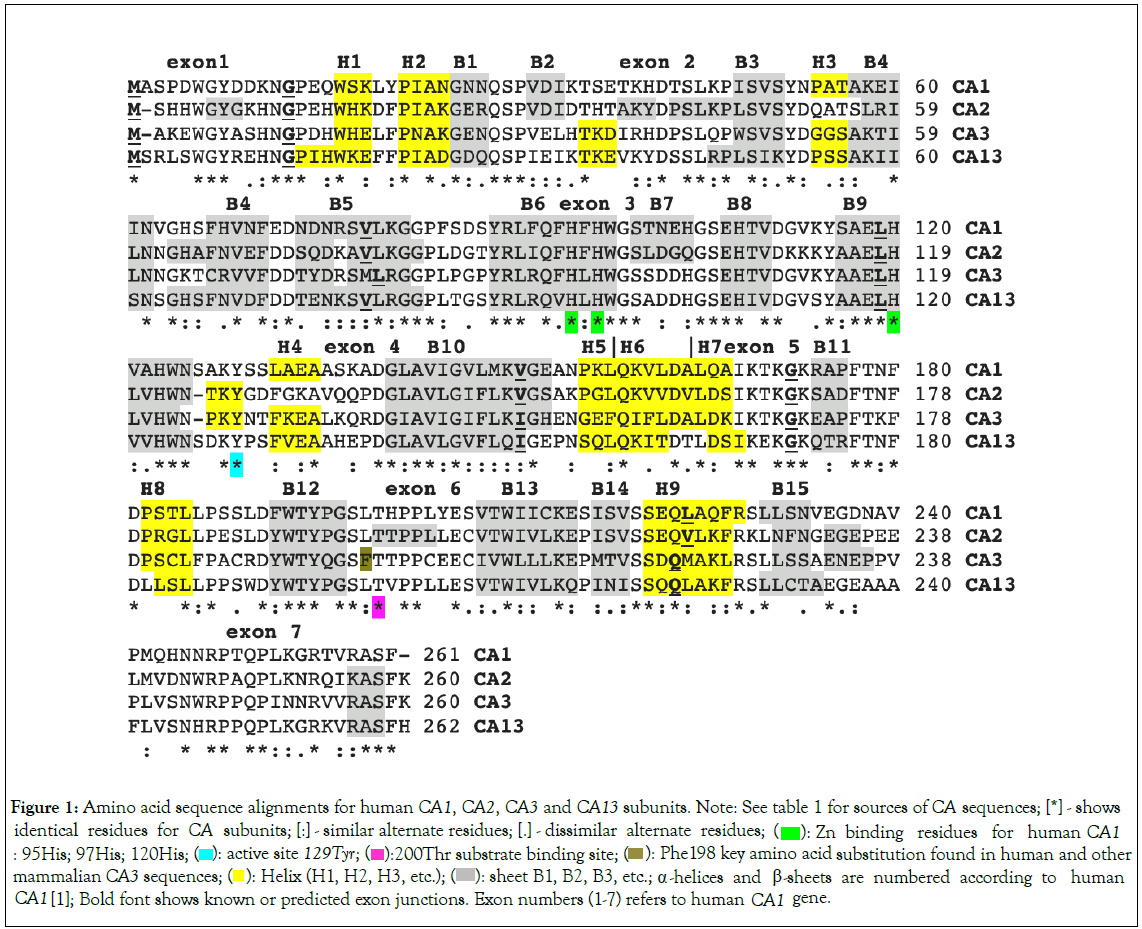
Figure 1: Amino acid sequence alignments for human CA1, CA2, CA3 and CA13 subunits. Note: See table 1 for sources of CA sequences; [*] - shows identical residues for CA subunits; [:] - similar alternate residues; [.] - dissimilar alternate residues;  Zn binding residues for human CA1 : 95His; 97His; 120His;
Zn binding residues for human CA1 : 95His; 97His; 120His;  Phe198 key amino acid substitution found in human and other mammalian CA3 sequences;
Phe198 key amino acid substitution found in human and other mammalian CA3 sequences;  sheet B1, B2, B3, etc.; α -helices and β-sheets are numbered according to human CA1 [1]; Bold font shows known or predicted exon junctions. Exon numbers (1-7) refers to human CA1 gene.
sheet B1, B2, B3, etc.; α -helices and β-sheets are numbered according to human CA1 [1]; Bold font shows known or predicted exon junctions. Exon numbers (1-7) refers to human CA1 gene.
X-ray crystallographic studies for human CA1 [1], CA2 [3], CA3 [5] and CA13 [8], have enabled the identification of key structural and catalytic residues among those aligned for these human CA sequences (Figure 1). The human CA1 sequence included Tyr129 which was identified as a catalytic residue; while His95, His97 and His120 were shown to be responsible for chelating the Zinc residue attheactivesite,whereas230Thr was involved in substrate binding. These residues were conserved among the human CA1, CA2, CA3 and CA13 sequences. Secondary structures among these CA isozymes were similar with 15 ß-sheets and 9 alpha helices observed for the human CA1 isozyme [1]. A key amino acid substitution was observed for human CA3, in comparison with the other isozymes, with respect to Phe198 (Leu in this position for CA1, CA2 and CA13), which has been shown to result in a steric constriction in the active site, resulting in much lower catalytic activity for this enzyme [5].
Predicted gene locations, exon structures and tissue expression for mammalian CA1, CA2, CA3 and CA13 genes
Table 1 and Figure 1, summarize the predicted locations and exon structures for CA1, CA2, CA3 and CA13 genes based upon BLAT interrogations of several mammalian and a bird genome using the sequences for the corresponding human CA1, CA2, CA3 and CA13 subunits (Table 1), and the UC Santa Cruz Web Browser [32]. These mammalian CA genes contained 7 coding exons with the predicted exon start sites in identical or similar positions (Figure 1). Figure 2, describes the tissue expression profiles for the human CA1, CA2, CA3 and CA13 genes and enzymes [31]. Human CA1 was predominantly expressed in colon and red cells, consistent with previous reports [2,4]. Human CA2 has a broader tissue expression profile, with highest expression levels being observed in the colon and stomach, but with significant expression in most tissues of the body, including the brain, red cells and kidney cortex. Human CA3 is almost exclusively expressed at high levels in skeletal muscle, as previously reported [5-7], exhibiting the highest CA expression levels among all human tissues examined. In contrast, tissue expression levels for CA13 were much lower as compared with the other CA isozymes (2-18 times), and with a broad distribution profile.
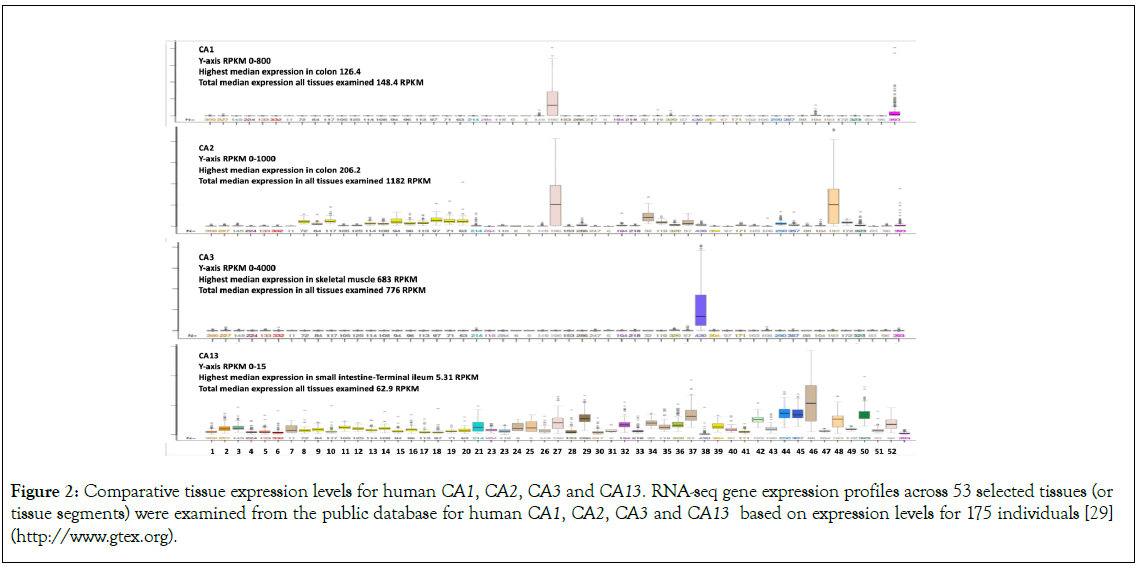
Figure 2: Comparative tissue expression levels for human CA1, CA2, CA3 and CA13. RNA-seq gene expression profiles across 53 selected tissues (or tissue segments) were examined from the public database for human CA1, CA2, CA3 and CA13 based on expression levels for 175 individuals [29] (http://www.gtex.org).
Figure 3, presents the predicted structures of human CA1, CA2 and CA13/CA3 gene transcripts [27]. There were 7 coding exons for the CA2 precursor mRNA sequence which contained several transcription factor binding sites in the 5’ region: SMARCA4 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4), which regulates transcription of genes by chromatin remodelling [34]; TFAP2C (transcription factor AP-2 gamma), which interacts with cellular enhancer elements to regulate transcription, particularly during early development [35]; and GATA3, a transcription factor and member of the zinc-finger regulatory proteins [36]. SMARCA4 was also located in the 5’ region of the human CA1, CA13 and CA3 genes which may indicate a similar role for this transcription factor for each of these genes. There were also 7 coding exons for the other CA genes examined, although the web site used to examine precursor mRNA structures for CA13 and CA3 suggested that these genes were contiguous in sequence [27]. CpG islands were observed for each of the 5’ regions for CA2 (CpG124); CA13 (CpG39); and CA3 (CpG33), which have the potential to undergo heritable epigenetic modification by methylation which can alter gene expression for these genes [37].

Figure 3: Gene structures and major isoforms for human CA1, CA2, CA3 and CA13 genes. Derived from AceView website: http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ [27]. Note: Mature isoform variants are shown with capped 5’- and 3’- ends for the predicted mRNA sequences. Exons are in solid colour; 5’- and 3’- untranslated regions of the genes are shown as open boxes; introns are shown as a line; 5’ → 3’ transcription directions, CpG islands and transcription factor binding sites are shown; mRNA isoforms for CA13 and CA3 are represented as being contiguous due to their proximal locations.
Functions of mammalianCA1, CA2, CA3 and CA13 families
Studies of mammalian CA1 and CA2 have supported their roles in red cells in the conversion of carbon dioxide to carbonate and bicarbonate ions, and in carbon dioxide export from the lungs, by catalysing the reverse reaction, converting bicarbonate ions back to carbon dioxide [2,9]. The proximal tubule of the kidney is predominantly responsible for bicarbonate transport from the kidney, especially involving CA2 activity in human and rodent kidneys [38]. This is supported by studies examining the impact of CA2 deficiency in mice which results in a urinary concentration defect [39]. Figure 2, demonstrates that high levels of CA2 expression are observed in human colon and stomach, which is consistent with a previous report supporting major roles for these enzymes in the gastrointestinal tract, including the production of gastric acid, bile, saliva and pancreatic juice, the absorption of salt and water in the GI tract and in facilitating intestinal electrolyte transport [40-42]. CA2 has been shown to form a transport metabolon with the electrogenic sodium-bicarbonate cotransporter (NBCe1), enhancing the bicarbonate transport capacity within kidney tubules [43]. Liver CA2 expression levels have been used as a molecular marker examining the therapy and diagnosis of hepatocellular carcinoma, with high CA2 expression levels being associated with overall survival and positive clinical treatment [44]. Bicarbonate ions have also been described as key factors in the regulation of sperm motility, and high concentrations of bicarbonate ions are present in the female genital tract, inducing an increase in sperm motility. Moreover, CA2 is distributed within the epididymis tract supporting sperm activity and assisting in fertilization [45]. Erythrocyte CAI deficiency has no major physiological impact, although CAII deficiency in other tissues may result in osteoporosis, renal tubular acidosis and brain calcification [46].
CA3 is highly expressed in muscle, particularly red skeletal muscles [47], with CA3 expression showing the highest level of expression as compared with all other human tissues examined for any of the CA isozymes (Figure 2). CA3 expression commenced early in neonatal mice and served as a marker of myogenesis, but is first detected in the myotomes of somites, before being restricted to developing slow muscle fibres [48]. In rats, CA3 is highly expressed in slow twitch skeletal muscle, adipocytes and liver, with lower levels detected in heart, prostate, kidney, brain and erythrocytes [6,49]. CA3 deficiency in skeletal muscles seems to play an important role in the pathogenesis of myasthenia gravis [50], causing muscle weakness [51]. Moreover, CA3 may play an important role as an antioxidant and protective agent, with the high levels of CA3 protein in skeletal muscle and liver acting as a reservoir of S-glutathione, through reversible binding to CA3 Cys188 [52]. In addition, transgenic expression of CA3 in mouse cardiac muscle appears to provide a mechanism for tolerating acidosis [53].
In contrast to CA1, CA2 and CA3, CA13 expression is uniformly low for all human tissues examined and appears to be performing a housekeeping function (Figure 2), as compared with the very high expression levels in stomach and red cells (CA1); colon and stomach (CA2) and skeletal muscle (CA3). CA13 expression is downregulated in colorectal cancer, together with CA1 and CA2 expression, which may reflect a level of coordination for the gene regulation for genes located within the CA gene complex [54].
Evolution of mammalian CA1, CA2, CA3 and CA13 gene families
Figure 4, presents a phylogenetic analysis of eutherian and marsupial mammalian CA1, CA2, CA3 and CA13 sequences, together with sequences from a bird species (brown kiwi) (Apteryx rowi). The phylogenetic tree supported a proposal for a sequence of gene duplication events, arising from an ancestral vertebrate CA2 gene, generating initially the CA3 gene, which is retained throughout subsequent vertebrate evolution, which is subsequently duplicated to form the CA1 and CA13 genes, both of which are retained throughout mammalian evolution. It appears that the CA2 gene is of ancient origin, with subsequent gene duplication events generating the CA1, CA3 and CA13 duplicated genes, all closely located within a CA gene complex located on human chromosome 8 or mouse chromosome 3. The ancient nature of CA2 among early vertebrates has been independently supported [55,56]. The CA13, CA1, CA3, CA2 gene complex is replicated among other eutherian and marsupial genomes, on chromosome 8 (human, rhesus monkey and green monkey genomes); chromosome 3 (mouse and opossum); chromosome 14 (cow); and on a brown kiwi chromosome segment, designated as NW_0140049943v1. It is apparent that the gene complex has been ‘flipped’ in the cow genome, with a reverse order of transcription, as compared with other eutherian genomes studies (Table 1) [57]. It is likely that close linkage for these genes is a product of the evolutionary events generating these CA genes, with selection potentially playing a role for retaining closely linked genes on mammalian and bird genomes, during>150 million years of evolution [58].

Figure 4: Phylogenetic tree of mammalian and bird CA1, CA2, CA3 and CA13 sequences. Note: A) The tree is labelled with the CA gene name and the name of the mammal or bird (brown kiwi); note the 4 major clusters for the CA1, CA2, CA3 and CA13 enzymes; gene duplication events generating the mammalian CA1, CA2, CA3 and CA13 gene families are proposed to have occurred in a CA2 ancestral gene leading to the formation of the marsupial and eutherian mammal groups. A genetic distance scale is shown. The number of times a clade (sequences common to a node or branch) occurred in the bootstrap replicates are shown. Only replicate values of 0.9 or more which are highly significant are shown. 100 bootstrap replicates were performed in each case. Sequences were derived from those reported in Table 1; B) Shows a representation of the primordial CA2 gene duplication events generating the CA2→CA3→CA1/CA13 genes within mammalian genomes.
Conclusion
The results of this study supported previous reports for 4 homologous CA genes and encoded cytoplasmic enzymes, CA1, CA2, CA3 and CA13, which are encoded by closely localized genes on human chromosome 8 and mouse chromosome 3 genomes. Knowledge of this CA1, CA2, CA3 and CA13 gene cluster is also reported in this paper for other eutherian and marsupial mammalian including rhesus monkey (Macacamulatta) and green (Chlorocebus sabeus) chromosome 8; cow (Bos taurus) chromosome 14; and opossum (Monodelphis domestica) chromosome 3. A similar CA1, CA2, CA3 and CA13 gene cluster was also observed in a New Zealand bird genome, the brown Kiwi (Apteryx mantelli). These genes are differentially expressed in human tissues, with very high expression levels observed for stomach and red cells (CA1); colon and stomach (CA1 and CA2); and skeletal muscle (CA3), whereas CA13 expression levels were much lower and more broadly distributed in human tissues. Phylogenetic studies of eutherian and marsupial mammalian CA1, CA2, CA3 and CA13 sequences, together with sequences from a bird species (brown kiwi) (Apteryx rowi), supported a proposal for a sequence of gene duplication events, arising from an ancestral vertebrate CA2 gene, generating initially the CA3 gene, which is retained throughout subsequent vertebrate evolution, which is subsequently duplicated to form the CA1 and CA13 genes, both of which are retained throughout mammalian evolution.
Acknowledgment
The advice of Dr Laura Cox of the Centre for Precision Medicine, Wake Forest School of Medicine, Winston Salem NC USA is gratefully acknowledged.
Conflict of Interest
The author reports no conflicts of interest.
References
- Kannan V, Kannan KK. Enzyme-structure interactions: Structure of human carbonic anhydrase 1 complexed with bicarbonate. J Mol Biol. 1994;241(2):226-232.
- Geers C, Gros G. Carbon dioxide transport and carbonic anhydrase in blood and muscle. Physiol Revs. 2000;80(2): 681-715.
[Crossref] [Google Scholar] [PubMed]
- Nair SK, Calderone TL, Christianson DW, Fierke CA. Altering the mouth of a hydrophobic pocket. Structure and kinetics of human carbonic anhydrase II mutants at residue Val-121. J Biol Chem. 1991;266:17320-17325.
[Crossref] [Google Scholar] [PubMed]
- Bekku S, Mochizuki H, Takayama E, Shinomiya E, Fukamachi H, Ichinose M, et al. Carbonic anhydrase 1 and 11 as a differentiation marker of human and rat enterocytes. Res Exp Med. 1998;1984:175-185.
[Crossref] [Google Scholar] [PubMed]
- Duda DM, Tu C, Fisher SZ, An H, Yoshioka C, Govindasamy L, et al. Human carbonic anhydrase III : Structural and kinetic study of catalysis and proton transfer. Biochem. 2005;44:10046-10053.
[Crossref] [Google Scholar] [PubMed]
- Harju A-K, Bootorabi F, Kuuslahti M, Supuran CT, Parkkila S. Carbonic anhydrase III: A neglected isozyme is stepping into the limelight. J Enz Inhib Med Chem. 2012;28(2):231-239.
[Crossref] [Google Scholar] [PubMed]
- Lehonen J, Shen B, Vihinen M, Casini A, Scozzafava A, Supuran CT, et al. Characterization of CA XIII, a member of the carbonic anhydrase isozyme family. J Biol Chem. 2004;279(4):2719-2727.
[Crossref] [Google Scholar] [PubMed]
- DiFiore A, Monti SM, Hilvo M, Parkkila S, Romano V, Scaloni A, et al. Crystal structure of the human carbonic anhydrase XIII in complex with acetazolamide. Proteins. 2009;74:164-175.
[Crossref] [Google Scholar] [PubMed]
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J. 2016;473(14):2023-2032.
[Crossref] [Google Scholar] [PubMed]
- Lomelino CL, Andring JT, McKenna R. Crystallography and its impact on carbonic anhydrase research. Int J Med Chem. 2018:9419521.
[Google Scholar] [PubMed]
- Ferraroni M, Tilli S, Briganti F, Chegwidden WR, Supuran CT, Wiebauer KE, et al. Crystal structure of a Zinc-activated variant of human carbonic anhydrase 1, CA1 Michigan 1: Evidence for a second Zinc binding site involving Arginine coordiation. Biochem.2002;41(20):6237-6244.
[Crossref] [Google Scholar] [PubMed]
- Supuran CT, Scozzafava A. Carbonic anhydrases as targets for medicinal chemistry. Bioorg Med Chem. 2007;15(13):4336-4350.
[Crossref] [Google Scholar] [PubMed]
- Yuan L, Wang M, Liu T, Lei Y, Miao Q, Li A, et al. Carbonic anhydrase 1-mediated calcification is associated with atherosclerosis, and methazolamide alleviates its pathogenesis. Front Pharmacol. 2019;109:766.
[Crossref] [Google Scholar] [PubMed]
- Eicher EM, Stern RH, Womack JE, Davisson MT, Roderick TH, Reynolds SC. Evolution of mammalian carbonic anhydrase loci by tandem duplication: Close linkage of Car-1 and Car-2 to the centromere region of chromosome 3 in the mouse. Biochem Genet. 1976;14(7-8):651-660.
[Crossref] [Google Scholar] [PubMed]
- Lowe N, Edwards YH, Edwards M, Butterworth PH. Physical mapping of the carbonic anhydrase gene cluster on chromosome 8. Genomics. 1991;10(4):882-888.
[Crossref] [Google Scholar] [PubMed]
- Edwards Y, Drummond F, Sowden J. Regulation of the CA1, CA2 and CA3 genes. The Carbonic Anhydrases: New Horizons. 2000:121-141.
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99(26):16899-16903.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-410.
[Crossref] [Google Scholar] [PubMed]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2003;12(6):994-1006.
[Crossref] [Google Scholar] [PubMed]
- Venter JC, Adams MD, Myers EW, Li PW, Murai RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1301-1351.
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316(5822):222-234.
[Google Scholar] [PubMed]
- Warren WC, Jasinska AJ, Garcia-Perez R, Svardal H, Tomlinson C, Rocchi M, et al. The genome of the vervet (Chlorocebus aethiops sabeus). Genome Res. 2015;25(12):1921-1933.
[Google Scholar] [PubMed]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002:420(6915):520-562.
[Crossref] [Google Scholar] [PubMed]
- Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, et al. A whole genome assembly of the domestic cow, Bos taurus. Genome Biology. 2009;10:R42.
[Crossref] [Google Scholar] [PubMed]
- Mikkelsen TS, Wakefield MJ, Aken B, Ameniya CT, Chang JL, Duke S, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167-177.
[Google Scholar] [PubMed]
- Le Duc D, Renaud G, Krishnan A, Almen MS, Huynen L, Prohaska SJ, et al. Kiwi genome provides insight into evolution of a nocturnal lifestyle. Genome Biol. 2015;16:147.
[Crossref] [Google Scholar] [PubMed]
- Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:1-4.
[Google Scholar] [PubMed]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497-3500.
[Crossref] [Google Scholar] [PubMed]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45(6):580-585.
[Google Scholar] [PubMed]
- Elango N, Yi SV. Functional relevance of pG island length for regulation of gene expression. Genetics. 2011:187(4):1077-1083.
[Crossref] [Google Scholar] [PubMed]
- Lesurf R, Cotto KC, Wang G, Griffith M, Kasaian K, Jones SJ, et al. ORegAnno 3.0: A community-driven resource for curated regulatory annotation. Nucleic Acids Res. 2016;44(D1):D126-D132.
[Crossref] [Google Scholar] [PubMed]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006.
[Google Scholar] [PubMed]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465-W469.
[Google Scholar] [PubMed]
- Hang CT, Yang J, Han P, Cheng H-L, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466(7302):62-67.
[Crossref] [Google Scholar] [PubMed]
- Chen D, Liu W, Zimmerman J, Pastor WA, Kim R, Hosohama L, et al. The TFAP2C- regulated OCT4 naïve enhancer is involved in human germline formation. Cell Rep. 2018;25(13):3591-3602.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35(3):337-348.
[Crossref] [Google Scholar] [PubMed]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10(5):295-304.
[Crossref] [Google Scholar] [PubMed]
- Purkerson JM, Schwartz GJ. The role of carbonic anhydrases in renal physiology. Kidney Int. 2007;71(2):103-115.
[Crossref] [Google Scholar] [PubMed]
- Sly WS, Whyte MP, Sundaram V, Tashian RE, Hewett-Emmett D, Guibaud P, et al. Carbonic anhydrase II deficiency in 12 families with the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. New Eng J Med. 1985;313(3):139-145.
[Crossref] [Google Scholar] [PubMed]
- Krishnan D, Pan W, Beggs MR, Trepiccione F, Chambrey R, Eladari D, et al. Deficiency of carbonic anhydrase II results in a urinary concentrating defect. Front Physiol. 2018;8:1108.
[Crossref] [Google Scholar] [PubMed]
- Lonnerholm G, Selking O, Winstrand PJ. Amount and distribution of carbonic anhydrases I and II in the gastrointestinal tract. Gastroenterology. 1985;88(5):1151-1161.
[Crossref] [Google Scholar] [PubMed]
- Charney AN, Wagner JD, Birnhaum GJ, Johnstone JN. Functional role of carbonic anhydrase in intestinal electrolyte transport. Am J Physiol. 1986;251(5):G682-G687.
[Crossref] [Google Scholar] [PubMed]
- Kivela AJ, Kivela J, Saamio J, Parkkilo S. Carbonic anhydrases in normal gastrointestinal tract and gastrointestinal tumours. World J Gastroenterol. 2005;11(2):115-163.
[Crossref] [Google Scholar] [PubMed]
- Becker HM, Deitmer JW. Carbonic anhydrase II increases the activity of the human electrogenic Na+/HCO3- cotransporter. J Biol Chem. 2007;282(18):13508-13521.
[Google Scholar] [PubMed]
- Zhang H, Zhuo C, Zhou D, Zhang F, Chen M, Xu S, et al. Association between the expression of carbonic anhydrase II and clinicopathological features of hepatocellular carcinoma. Oncol. Lett. 2019;17(6):5721-5728.
[Crossref] [Google Scholar] [PubMed]
- Wandernoth PM, Mannowetz N, Szczyrba J, Grannemann L, Wolf A, Becker HM, et al. Normal fertility requires the expression of carbonic anhydrases II and IV in sperm. J Biol Chem. 2015;290(49):29202-29216.
[Crossref] [Google Scholar] [PubMed]
- Borthwick K, Kandemir N, Topaloglu R, Kornak U, Bakkaloglu AY, Yordam NU, et al. A phenocopy of CAII deficiency: A novel genetic explanation for inherited infantile osteopetrosis with distal renal tubular acidosis. J. Med. Genet. 2003;40(2),115-121.
[Crossref] [Google Scholar] [PubMed]
- Holmes RS. A comparative electrophoretic analysis of mammalian carbonic anhydrase isozymes: Evidence for a third isozyme in red skeletal muscles. Comp Biochem Physiol. 1976;578:117-120.
[Crossref] [Google Scholar] [PubMed]
- Lyons GE, Buckingham ME, Tweedie S, Edwards YH. Carbonic anhydrase III, an early mesodermal marker, is expressed in embryonic mouse skeletal muscle and notochord. Biochem J. 1991;111:233-244.
[Crossref] [Google Scholar] [PubMed]
- Stanton LW, Ponte PA, Coleman RT, Snyder MA. Expression of CAIII in rodent models of obesity. Mol Endocrinol. 1991;5:860-866.
- Du A, Huang S, Zhao X, Feng K, Zhang S, Huang J, et al. Suppression of CHRN endocytosis by carbonic anhydrase CAR3 in the pathogenesis of myasthenia gravis. Autophagy. 2017;13(11):1981-1994.
[Crossref] [Google Scholar] [PubMed]
- Trouth AJ, Dabi A, Solieman N, Kurukumbi M, Kalyanam J. Myasthenia gravis: A review. Autoimmune Dis. 2012:874680.
- Di Fiore A, Monti DM, Scaloni A, De Simone G, Monto SM. Protective role of carbonic anhydrases III and VII in cellular defence mechanisms upon redox balance. Oxid Med Cell Longev. 2018:2018306.
[Crossref] [Google Scholar] [PubMed]
- Feng HZ, Jin JP. Transgenic expression of carbonic anhydrase III in cardiac muscle demonstrates a mechanism to tolerate acidosis. Am J Physiol. Cell Physiol. 2019;317(5):C922-C931.
[Crossref] [Google Scholar] [PubMed]
- Kummola L, Hamalainen JM, Kivela AJ, Saarnio J, Karttunen T, Parkkila. Expression of a novel carbonic anhydrase, CAXIII, in normal and neoplastic colorectal mucosa. BMC cancer. 2005; 5:41.
[Crossref] [Google Scholar] [PubMed]
- Lund SG, Dyment P, Gervais MR, Moyes CD, Tufts BL. Characterization of erythrocyte carbonic anhydrase in an ancient fish, the longnose gar (Lepisosteus osseus). J Comp Physiol. 2022;B172(6):467-476.
[Crossref] [Google Scholar] [PubMed]
- Esbaugh AJ, Tufts BL. Tribute to R. G. Boutilier: Evidence of a high activity carbonic anhydrase isozyme in the red cells of an ancient vertebrate, the sea lamprey Petromyzon marinus. J Exp Biol. 2006;209(7):1169-1178.
[Crossref] [Google Scholar] [PubMed]
- Brusatte SL, O’Connor JK, Jarvis ED. The origin and diversification of birds. Curr. Biol. 2015;25(19):R888-R898.
[Crossref] [Google Scholar] [PubMed]
Citation: Holmes RS (2024). Review: Comparative Studies and Evolution of Mammalian and Bird CA1, CA2, CA3 and CA13 Genes and Proteins. J Data Mining Genomics Proteomics. 15:338.
Copyright: © 2024 Holmes RS. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

