Indexed In
- Online Access to Research in the Environment (OARE)
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- Scimago
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- University Grants Commission
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 14, Issue 1
Comparative Analysis of Genetic Diversity in Natural Population of Labeo rohita and Cirrhinus marigala by Using Species Specific Molecular Markers
Faisal Tasleem1*, Shahid M1, Hina Fatima2 and Numan Gulzar M22Department of Biochemistry, University of Agriculture, Faisalabad, Pakistan
Received: 23-Jul-2022, Manuscript No. JARD-22-17546; Editor assigned: 25-Jul-2022, Pre QC No. JARD-22-17546 (PQ); Reviewed: 08-Aug-2022, QC No. JARD-22-17546; Revised: 21-Sep-2022, Manuscript No. JARD-22-17546 (R); Published: 28-Sep-2022, DOI: 10.35248/2155-9546.22.13.718
Abstract
Labeo rohita, commonly known as rohu and Cirrhinus marigala, popularly known as morakkha are economically most important and extensively cultured fish species in the entire Indian subcontinent. Genetic evaluation of these species is necessary for their proper supervision conservation and profitable production. Various types of molecular markers are available but SSR are most important one due to their equal distribution, abundance in the genome, co- dominance, polymorphic, low cost detection and multi-allelic nature. In this study ten samples of each species were collected from Head Punjnad, Head Muhammad Wala and Head Trimmu region of river Chenab, Pakistan. After the extraction of DNA, amplification of five simple sequence repeat markers, their resolution on PAGE and allelic scoring for the genotypic data was used for the analysis of different genetic diversity indices with the help of POPGENE version 1.32, FSTAT version 2.9.3.2, Jaccard and Dice similarity coefficient, simple matching analysis and SIMQUAL program of NTSYS-PC package.
In case of Labeo rohita, total 26 alleles with 260 loci were detected out of which 189 were found polymorphic. Polymorphism for the five markers varies from 43.33%-96% with mean value of 72.69%. Allelic frequency ranges from 0.2000-0.9000 with 0.4600 mean value, allele numbers varies from 4.000-9.000 with average value of 5.2000, gene diversity varies from 0.1800-8800 with mean value of 0.6360 and PIC value ranges from 0.1638-0.8680 with 0.6012 mean value. Pair wise genetic difference among ten collected samples varies from 0.400-0.900 while pair wise genetic similarity ranges from 0.100-1.000. Cluster analysis based on UPGMA divided ten samples of Labeo rohita into three clusters and three sub clusters. Similarly, five SSR markers of Cirrhinus marigala show 30 alleles with 300 loci across ten samples with 240 polymorphic loci. Polymorphism ranges from 70%-96% with average value of 80%. Allelic frequency varies from 0.4000-0.9000 with average value of 0.6600. Allele numbers varies from 4.000-9.000 with 6.000 as mean value. Gene diversity varies from 0.1800-0.7800 with mean value of 0.4680. PIC value ranges from 0.1638-0.7578 with mean value of 0.4422. Pair wise genetic difference among ten samples of Cirrhinus marigala ranges from 0.2000-0.800 and pair wise similarity ranges from 0.200-1.000. Cluster analysis based on UPGMA divided all the samples of Cirrhinus marigala into three clusters and three sub clusters. This study reveals that Government of Pakistan and interested agencies should take serious steps for the improvement of genetic structure of these species especially Cirrhinus marigala.
Keywords
Labeo rohita; Cirrhinus marigala; Genetic diversity; Simple sequence repeat markers
Introduction
Fish is an excellent source of good quality meat, vitamins, poly unsaturated fatty acids and minerals. Fish meat is easily digestible. It is a renewable resource of income and nutrition for a country [1]. Fisheries sector provides jobs and investment opportunities to lot of concerned peoples [2]. Pakistan export fish and fish products to more than 50 countries especially European union countries, China, Malaysia, Japan, Singapore, Sri Lanka, Hong Kong, Saudi Arabia and S. Korea and earns 232.5 million Dollars annually [3]. More than 24,600 fish species are found in Pakistan [4].
Labeo rohita is known as rohu in Pakistan. In Latin, it is given the name of Labeo due to having large lips. It has arched head with silvered colored cyprinid shape body. It may gain 50 kg body weight with 2 meter body length. It is omnivorous and breeds during moon soon season [5].
Similarly, Cirrhinus marigala is commonly known as morakkha and naini in Pakistan. This is also member of Indian major carps which belongs to genus Cirrhinus. The body of this specie is somewhat compressed, oblong and large with round nose, broad mouth, sharp lower lips and fringed upper lips [6]. On the palate there are various fingers like projection.
Indus, Chenab, Jhelum, Ravi, Beas and Sutlej are more famous rivers of Pakistan providing superlative vegetative, nutritive and climatic zones to lot of fish species [7]. Instead of tremendous scope, potential and earning through fishery sector, studies on genetic improvements and managemental practices are not found in literature. This is the reason that this study was designed for the evaluation of genetic potential and genetic variations of two (Cirrhinus marigala and Labeo rohita) species.
Both these species (Labeo rohita and Cirrhinus marigla) are well known table fish of South East Asians countries especially in Pakistan, India and Bangladesh [8]. This is the reason that these species are commonly cultured in the private and public hatcheries of these countries [9]. Labeo rohita is one of the top ten fish species being cultured worldwide [10]. In order to meet the fish meat requirement of human population, we should enhance the breeding and rearing programs of these species. In Pakistan, about 99% seed of Labeo rohita is provided by public and private hatcheries [11].
There are many factors such as diseases, agricultural pollutants, overfishing, poor quality brooder seed, Dam’s construction, inbreeding, inappropriate management etc. which affects the growth and survival of fresh water species very badly [12]. Ii is necessary to overcome these factors for the highest yield of fish. By understanding the genetic makeup of fishes, provision of better guidelines to farmers and via better policies we can enhance the production of fishes. According to the conservation biologist, in altering environment, restocking and genetic variations are major factors for controlling the fish production [13].
Living organisms ensure there survivability by passing through various evolutionary process [14]. Genetic variation is one of the evolutionary steps which enhance the survivability of living things even in changing environments [15]. Population genetics is helpful for understanding the evolutionary process of fish [16].
Genetic study of any species can be conducted by using different types of genetic or molecular markers. Simple Sequence Repeat (SSR) are one of the molecular markers which are widely used in genetic study due to their multi-allelic nature, co-dominance, relative abundance, hyper variability, relative abundance, reproducibility, high throughput genotyping and fixed location on the chromosomes [17]. Due to having multiple allelic natures, SSR markers reveal exact genotypic variations between samples of even same specie [18]. On the basis of such properties, species specific SSR markers have been used in this study to check the extent of genetic variations among the individuals of Labeo rohita and Cirrhinus marigala.
Materials and Methods
Sampling of fish for DNA extraction
Ten samples of Cirrhinus marigla and ten samples of Labeo rohita were collected from three areas (Head Trimmu, Head Muhammad Wala and Head Punjnad) of river Chenab.
The detail of Cirrhinus marigala and Labeo rohita samples codes along with their collection sites have been summarized in Tables 1 and 2 respectively. After catching the fish samples from running water of river Chenab with the help of net, they were packed in zip lock polythene bags and transferred to the University Lab the on the same day where they were stored at -20˚C in the freezer.
| Serial number | Sample code | Sample type | Name of collection site |
|---|---|---|---|
| 1-4 | CHPND1,CHPND2,CHPND3,CHPND4 | Muscle | Head Punjnad |
| 5-7 | CHMW5, CHMW6, CHMW7 | Muscle | Head Muhammad Wala |
| 8-10 | CHT8,CHT9,CHT10 | Muscle | Head Trimmu |
CHPND (1-4): Cirrhinus marigala collected from Head Punjnad, CHMW (5-7): Cirrhinus marigala from Head Muhammad wala,CHT (8-10): Cirrhinus marigala from Head Trimmu
Table 1: Sample codes of Cirrhinus marigala along their site of collections.
| Serial number | Sample code | Sample type | Sample source |
|---|---|---|---|
| 1-4 | LHPND1, LHPND2, LHPND3, LHPND4 | Muscle | Head punjnad |
| 5-7 | LHMW5, LHMW6, LHMW7 | Muscle | Head Muhammad Wala |
| 8-10 | LHT8, LHT9, LHT10 | Muscle | Head Trimmu |
| LHPND (1-4): Labeo rohita collected from Head Punjnad, LHMW (5-7): L. rohita from Head Muhammad wala, LHT (8-10): L. rohita from Head Trimmu | |||
Table 2: Sample codes of Labeo rohita with their sites of collection.
DNA extraction
DNA from the dorsal muscle of each fish sample was extracted with slight modification in Natalia Bello, 2001 method. In this method a 2 cm-3 cm piece of meat was excised and chopped with surgical blade. After digestion of meat with the help of lyses buffer and proteinase K, precipitation of DNA by Phenol: Chloroform: Isoamylalcohol and purification of DNA by chilled ethanol and 2M NaCl solution was done. The quality of DNA was checked by gel electrophoresis under UV Trans illuminator and quantity of DNA was checked by spectrophotometer (Perkin Elmer Ltd. UK).
Polymerase Chain Reaction (PCR)
20 ul PCR reaction mixture containing 10.75 ul PCR water, 2 ul template DNA, 2.5 ul dNTPs, 2.5 ul 10X buffer, 1 ul forward, 1 ul reverse primer and 0.25 ul Taq DNA polymerase was used for the amplification used DNA with the help of Gene Amp system (Applied Biosystems). PCR profile was set as; 1 cycle of initial denaturation at 95°C for 5 minute, 35 cycles of final denaturation at 94°C for 1 minute, annealing for 1 minute at 55°C-60°C (varies from primer to primer), initial and final extention at 72°C for 2 and 7 minutes (Tables 3 and 4).
| Sr.No. | Primer name | Accession no. | Sequence 5’-3’ | Annealing temp. | GC contents |
|---|---|---|---|---|---|
| 1 | Clone Lr33 | AM269523 | F: 5′-CTT GCC GCT GTC TTT CGC-3′ (18mer) | 59.85°C | 61.11% |
| R: 5′-GCC ACT GTT TAG CTT CAC AGG-3′ (21mer) | 52.38% | ||||
| 2 | Clone Lr30 | AM231179 | F: 5′-CAT ACA CGC CGA CCT CCC-3′ (18mer) | 60.7°C | 66.67% |
| R: 5′-CCA GGC CTC TGT GCT TCC-3′ (18mer) | 66.67% | ||||
| 3 | Clone22 | AM285342 | F: 5′-TCT GTG TGT GTG TGT GCG-3′ (18mer) | 57.25°C | 55.56% |
| R: 5′-ATG TGG AGG AAT GCC GGC-3′ (18mer) | 61.11% | ||||
| 4 | CLONE Lr38 | AM269528 | F: 5′-AGC TGT GCG ATT GCC CAT-3′ (18mer) | 57.25°C | 55.56% |
| R: 5′-GGT TTG GAA GCG CTC CCA-3′ (18mer) | 61.11% | ||||
| 5 | Clone Lr32 | AM23118 | F: 5′-GGC TCT CAG AAG ACC AGC G-3′ (19mer) | 60.05°C | 63.16% |
| R: 5′-TCC CCT GCC GTT CTC TGA-3′ (18mer) | 61.11% |
Table 3: SSR primers information for Labeo rohita.
| Sr. No. | Name | Sequence 5′-3′ | Annealing tem (°C) |
|---|---|---|---|
| 1 | C. marigala_A(F) | CCA TAA GGT AAA GCG CTG GC | 57 |
| C. marigala_A(R) | ACA AAG GTG TGT GTG TTG TGT G | ||
| 2 | C. marigala_D(F) | CAC ACA CGT TAA AAC ACA CGC | 58.4 |
| C. marigala_D(R) | CTG AGC GAA ACT GCA CAA GC | ||
| 3 | C. marigala_F(F) | CAG CGC GCA CAC AGA GA | 56 |
| C. marigala_F(R) | CTT TCG GCG AAT GGG CTG | ||
| 4 | C. marigala_I(F) | CCA TAA GGT AAA GCG CTG GC | 57 |
| C. marigala_I(R) | AAA GGT GTG TGT GTT GTG TGT G | ||
| 5 | C. marigala_L(F) | CAG CGC GCA CAC AGA GAG | 56.5 |
| C. marigala_L(R) | CTT TCG GCG AAT GGG CTG |
Table 4: SSR primers information for Cirrhinus marigala.
In this study, five SSR markers for Labeo rohita and five for Cirrhinus marigala was used which were designed and purchased from the Humanizing Genomics Macrogen Company. The detail of each primer is given below [19].
Polyacrylamide Gel Electrophoresis (PAGE)
30% polyacrylamide gel containing 11.25 ml of 30% acrylamide, 400 ul APS, 26.25 ml 1X TBE buffer, 30 ul TEMED and 400 ul APS solution was used in horizontal PAGE apparatus (Cat#MGV-220-33 CBS scientific UK) for the separation and detection of PCR amplified products. This apparatus was given current of 100 A and 200 voltage for 80 minutes. Then the gel was passed through three steps (fixation, staining and developing) of silver staining and at the end it was seen under UV trans illuminator. Make the picture of visible bands, score and apply software for genetic analysis.
Software and analytical packages
An excel sheet was made on the basis of scoring which ultimately subjected to statistical analysis POPGENE version 1.32 and FSTAT version 2.9.3.2 for measurement of allele numbers, allelic frequency, Polymorphism Information Content (PIC) and gene diversity values for each primer [20,21]. Jaccard and Dice similarity coefficient and simple matching analysis was used for the calculation of similarity index and frequency based Genetic difference was calculated by using NTSYS_PC package statistical software [22,23]. Cluster of all the samples were made by using Un-weighted Pair Group Method with Arithmetic Mean (UPGMA) in SAHN program of NTSYS PC version 2.11 J Package.
Results
Genetic diversity among the Labeo rohita and Cirrhinus marigala samples of river Chenab was evaluated by using five simple sequence repeat markers for each species. The results of genetic parameters after applying statistical analysis on the scoring sheet are shown in Table 5.
| Marker | Major allele frequency | Allele No. | Gene diversity | PIC | Polymorphism |
|---|---|---|---|---|---|
| CloneLr22 | 0.3000 | 4.0000 | 0.7400 | 0.6918 | 82.5% |
| Lr30 | 0.9000 | 5.0000 | 0.1800 | 0.1638 | 96% |
| Lr32 | 0.5000 | 4.0000 | 0.6400 | 0.5812 | 95% |
| Lr33 | 0.4000 | 4.0000 | 0.7400 | 0.7014 | 77.5% |
| Lr38 | 0.2000 | 9.0000 | 0.8800 | 0.8680 | 43.33% |
| Mean | 0.4600 | 5.2000 | 0.6360 | 0.6012 | 72.69% |
Table 5: Values of basic genetic parameters of Labeo rohita samples.
Five simple sequence repeat markers (clone Lr22, Lr30, Lr32, Lr33 and Lr38) were used for Labeo rohita samples. Different markers show different values of genetic parameters. Values of gene diversity varies from 0.1800 (Lr30) to 0.8800 (Lr38) and 0.6360 as an average value. Allele number ranges from 4.000-9.000 with 5.200 as a mean value. Values of allelic frequency varies from 0.2000 (Lr38) to 0.9000 (Lr30) with 0.4600 average value. Polymorphism information content values are best indicators as these values are proportional to genetic diversity. PIC values ranges from 0.1638 (Lr30) to 0.8680 (Lr38). All these values indicates moderate to high genetic variation between the samples of Labeo rohita specie.
Phylogenetic tree of Labeo rohita samples
Phylogenetic tree represents evolutionary relationship among the samples. UPGMA a distance based algorithm was used for the construction of phylogenetic tree. The resultant dendrogram divided ten individuals of L. rohita into three clusters and three sub clusters as shown in Figure 1.
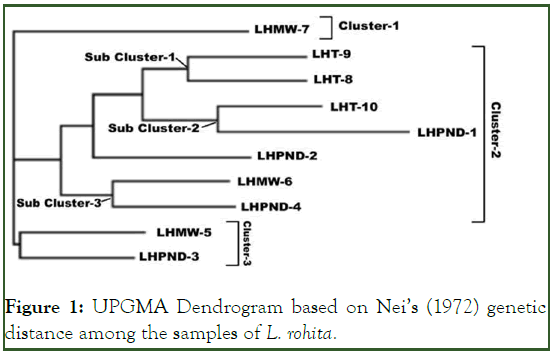
Figure 1: UPGMA Dendrogram based on Nei’s (1972) genetic distance among the samples of L. rohita.
Cluster 1 contains LHMW7 (Labeo rohita sample no.7 from Head Muhammad Wala) which shows pair wise genetic difference 0.600-0.900 from other samples. Sub cluster 1 consists of two samples of Head Trimmu. Sub cluster 2 consists of one sample of Head Trimmu while other of Head Punjnad. Sub cluster 3 consists of LHMW6 and LHPND4. Cluster 3 consists of LHMW5 and LHPND3. Pair wise genetic similarity between these samples is 0.6000. LHMW5 is sample of Head Muhammad Wala and LHPND4 is sample of Head PUNJNAD. Despite of geographical distribution, both samples falls in the same cluster.
LHPND 1-4 (Labeo rohita collected from Head Punjnad), LHMW 5-7 (Labeo rohita from Head Muhammad Wala, LHT 8-10 (Labeo rohita from Head Trimmu) (Table 6).
| Marker | Major allele frequency | Allele No. | Gene diversity | PIC | Polymorphism |
|---|---|---|---|---|---|
| Cirrhinus marigal (A) | 0.9000 | 4.0000 | 0.1800 | 0.1638 | 95% |
| Cirrhinus marigal (D) | 0.9000 | 5.0000 | 0.1800 | 0.1638 | 96% |
| Cirrhinus marigal (F) | 0.7000 | 5.0000 | 0.4800 | 0.4500 | 72% |
| Cirrhinus marigal (I) | 0.4000 | 7.0000 | 0.7200 | 0.6756 | 78.57% |
| Cirrhinus marigal (L) | 0.4000 | 9.0000 | 0.7800 | 0.7578 | 70% |
| Mean | 0.6600 | 6.0000 | 0.4680 | 0.4422 | 80% |
Table 6: Values of basic parameters of genetic diversity.
The values of genetic parameters in case of Cirrhinus marigala are different from those of Labeo rohita samples. All these parameters also show moderate to high genetic variations among the samples of Cirrhinus marigala.
Phylogenetic tree
A dendrogam based on UPGMA divided ten samples into three clusters and two sub clusters as shown in Figure 2. Cluster 1 consists of only sample no.9 (CHT9) which was collected from Head Trimmu. Cluster 2 consists of seven individuals (CHPND4, CHT10, CHMW7, CHMW6, CHPND1, CHT8 and CHPND2). Further it is divided into two sub clusters (sub cluster 1 and sub cluster 2). Sub cluster 1 consists of two samples. One sample (CHPND4) which was collected from Head Punjnad and second sample (CHT10) was collected from Head Trimmu. Sub cluster 2 is consists of CHMW6 and CHMW7 samples. Both these samples were collected from Head Muhammad Wala and show genetic difference (0.4000). The remaining individuals of cluster 2 are CHPND1, CHPND2 and CHT8. CHPND1 and CHPND2 were collected from Head Punjnad and CHT8 was collected from Head Trimmu. Cluster 3 consists of only two samples (CHPND3 and CHMW5) as shown in Figure 2.
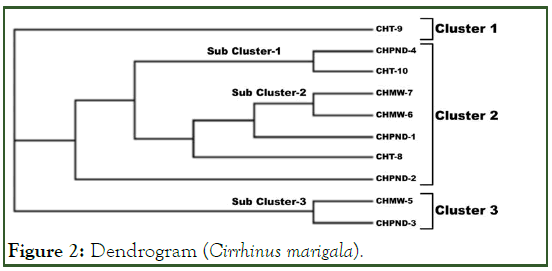
Figure 2: Dendrogram (Cirrhinus marigala).
[UPGMA Dendrogram based on Nei’s (1972) genetic distance among the samples of Cirrhinus marigala
CHPND 1-4 (Cirrhinus marigala samples of Head Punjnad), CHMW 5-7 (Cirrhinus marigala from Head Muhammad Wala), CHT 8-10 (Cirrhinus marigala from Head Trimmu).
Discussion
Knowledge of genetic diversity is helpful for effective management and preservative strategies [23]. Ten commercial fish species including Labeo rohita and Cirrhinus marigala of Asian region are genetically degrading gradually [25]. Therefore it is necessary to take serious steps for conservation and management of these species.
This study is conducted for the evaluation of genetic status of Indian major carps (Labeo rohita and Cirrhinus marigala) in the wild population of river Chenab, Pakistan. Species specific SSR markers proved to be very useful tools for this study. In this study numbers of alleles ranges from 4-9 in case of Labeo rohita and 2-7 for Cirrhinus marigala populations which are less as compared to other studies [26]. The reason for such limited numbers of alleles indicates the low level of genetic potential in the studied species of river Chenab. This may be due to using less numbers of brooders for mass creation of fish seed [11].
In this study Labeo rohita samples show less allelic frequency (0.2000-0.9000) with average value of 0.4600 as compared to samples of Cirrhinus marigala whose allelic frequency ranges from (0.4000-0.9000) with average value of 0.6600. The degree of genetic deviation within a population is known as Heterozygosity (H). Heterozygosity is resulted due to long time naturally adaptation to fluctuating environment [27]. Gene diversity is the measure of deviation of a gene at single locus per individuals. In case of Labeo rohita population, its value ranges from 0.1800-0.8800 with 0.6360 as an average value. These values are almost similar to the results obtained by who concluded that genetic diversity among Labeo rohita samples of six hatcheries varies from 0.661-0.717 [27]. But these values are much greater as compared to 0.05 as studied by [28]. Lowest gene diversity (0.1800) is shown by marker Lr30 and highest value (0.8800) is shown by Lr38. In case of Cirrhinus marigala these values ranges from 0.1800 (Cirrhinus marigal A, Cirrhinus marigala D) to 0.7800 (Cirrhinus marigala L) with an average value of 0.4680. It is clear from the results of this parameter that Labeo rohita population is genetically more diverse than Cirrhinus marigala population.
Similarly Polymorphism Information Content (PIC) is basic tools for evaluating the genetic potential of any species. In this study its value ranges from 0.1638-0.8680 for Labeo rohita samples. Lower value is shown by Lr30 and upper value is shown by Lr38. The mean value shown by all the primers of Labeo rohita samples is 0.6012. Such a highest PIC value is indication of greater genetic variations among the Labeo rohita samples. In case of Cirrhinus marigala PIC value ranges from 0.1638-0.7578 with average value of 0.4422. This average value is much less than the average value of PIC for Labeo rohita samples. PIC value for Ctenophyrynogodon idella with the help of SSR markers was obtained (PIC>0.7) in one of the study conducted in China by. Therefore we can say easily on the basis of these results that Labeo rohita species is more diverse as compared to Cirrhinus marigala species.
Pair wise genetic differentiation (FST) shows the degree of genetic variation between the two comparing sequences or individuals. In case of Labeo rohita population, its value ranges from 0.400 to 0.900. This is very highest value that indicates that Labeo rohita population of river Chenab is genetically more diverse. This population has adopted themselves according to the environmental conditions of this river. In case of Cirrhinus marigala the values of (FST) ranges from 0.2000-0.8000. Mostly pair of Cirrhinus marigala individuals shows the value of (FST) of 0.2000 and 0.4000. Comparatively these values are much less as compared to the values for Labeo rohita samples. According to the genetic distance between the Labeo rohita population of Barisal (Bangladesh) and Indian region was obtained 0.7221 [29]. On the basis of results of all these parameters, I can say that Labeo rohita population is genetically more diverse as compared to the population of Cirrhinus marigala.
Dendrogram was constructed for each species separately on the basis of UPGMA (unweighted pair group with arithmetic mean). This is distance based algorithms which separate the more diverse sequences away from each other’s. Closely related sequences or individuals are kept in the same cluster [30]. In this study ten samples of Labeo rohita fall in three clusters. Seven individuals fall in cluster 2 and remaining two samples make cluster 3. Similarly, the samples of C. marigala also make three clusters.
Conclusion
In this study simple sequence repeat markers proved best tools for the evaluation of genetic diversity within and among the population of Labeo rohita and Cirrhinus marigala. By comparing results of different genetic parameters, it is clear that Labeo rohita samples of river Chenab are genetically more diverse than those of Cirrhinus marigala. It means L. rohita is more adopted in the natural environment of river Chenab. This is the reason that L. rohita is much more preffered and cultivated species in Pakistan.
References
- Dwivedi AC, Nautiyal P. Stock assessment of fish species Labeo rohita, Tor tor and Labeo calbasu in the rivers of Vindhyan region, India. J Environ Biol. 2012;33(2):261-264. [Crossref]
- Danish M, Singh IJ. Genetic Diversity Analysis of Labeo rohita (Hamilton, 1822) From Hatchery and Dhaura Reservoir of Uttarakhand by Using Microsatellite Markers. Int J Curr Microbiol App Sci. 2017;6(6):1432-1442.
- Akhter N, Wu B, Memon AM, Mohsin M. Probiotics and prebiotics associated with aquaculture: A review. Fish Shellfish Immunol. 2015;45(2):733-741.
- Raja M, Malaiammal P, Nandagopal S, Muralidharan M, Arunachalam M. Comparative analysis of genetic diversity among three species of the freshwater fish genus Garra (Osteichthys: Cyprinidae) using restriction fragment length polymorphism. Int J Biosci. 2012;2(3):88-96.
- Alam M, Jahan M, Hossain M, Islam M. Population genetic structure of three major river populations of rohu, Labeo rohita (Cyprinidae: Cypriniformes) using microsatellite DNA markers. Gene Genom. 2009;31(1):43-51.
- Jhingran VC, Pullin RSV. A hatchery manual for common, Chinese and Indian major carps. ICLARM Stud Rev. 1988;11:191.
- Rafique M, Khan NU. Distribution and status of significant freshwater fishes of Pakistan. Rec Zool Surv Pak. 2012;21:90-95.
- Talwar PK, Jhingran AG. Inland fishes of India and adjacent countries. CRC Press, Florida, USA. 1991;19:135-136.
- Abbas K, Zhou X, Li Y, Gao Z, Wang W. Microsatellite diversity and population genetic structure of yellowcheek, Elopichthys bambusa (Cyprinidae) in the Yangtze River. Biochem Syst Ecol. 2010;38(4):806-812.
- Alam MA, Akanda MS, Khan MM, Alam MS. Comparison of genetic variability between a hatchery and a river population of rohu (Labeo rohita) by Allozyme Elctrophoresis. Pakistan J Biol Sci. 2002;5:959-961.
- Islam MS, Alam MS. Randomly amplified polymorphic DNA analysis of four different populations of the Indian major carp, Labeo rohita (Hamilton). J Appl Ichthyol. 2004;20(5):407-412.
- Gaston KJ. Global patterns in biodiversity. Nature. 2000;405(6783):220-227.
- Ponzoni RW, Nguyen NH, Khaw HL. Genetic improvement programs for aquaculture species in developing countries: Prospects and challenges. Proc Assoc Advmt Anim. Breed Genet. 2009;18:342-349.
- Colihueque N, Araneda C. Appearance traits in fish farming: progress from classical genetics to genomics, providing insight into current and potential genetic improvement. Front Genetica. 2014;5:251.
- Díaz S, Purvis A, Cornelissen JH, Mace GM, Donoghue MJ, Ewers RM, et al. Functional traits, the phylogeny of function and ecosystem service vulnerability. Ecol Evol. 2013;3(9):2958-2975.
- Waples RS, Pess GR, Beechie T. Evolutionary history of Pacific salmon in dynamic environments. Evol Appl. 2008;1(2):189-206.
- Wang L, Shi X, Su Y, Meng Z, Lin H. Loss of genetic diversity in the cultured stocks of the large yellow croaker, Larimichthys crocea, revealed by microsatellites. Int J Mol Sci 2012;13(5):5584-5597.
- Legesse BW, Myburg AA, Pixley KV, Botha AM. Genetic diversity of African maize inbred lines revealed by SSR markers. Hereditas. 2007;144(1):10-7.
- Patel A, Das P, Swain SK, Meher PK, Jayasankar P, Sarangi N. Development of 21 new microsatellite markers in Labeo rohita (rohu). Anim Genet. 2009;40(2):253-254.
- Yeh FC, Yang RC, Boyle TB, Ye ZH, Mao JX. POPGENE, the user-friendly shareware for population genetic analysis, molecular biology and biotechnology centre. University of Alberta, Canada. 1997;19:213-232.
- Goudet JF. FSTAT (version 1.2): a computer program to calculate F-statistics. J Heredity. 1995;86(6):485-486.
- Sokal RR. The principles and practice of numerical taxonomy. Taxon. 196312(5):190-199.
- Nei M. Genetic distance between populations. Am Nat. 1972;106(949):283-292.
- Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. Estimation of census and effective population sizes: The increasing usefulness of DNA-based approaches. Conserv Genet. 2010;11(2):355-373.
- Altaf N, Khan AR. Variation within kinnow (Citrus reticulata) and rough lemon (Citrus jambheri). Pak J Bot. 2008;40(2):589-598.
- Alam MS, Islam MS. Population genetic structure of Catla catla (Hamilton) revealed by microsatellite DNA markers. Aquac. 2005;246(1-4):151-60.
- Chapman LJ, Mckenzie DJ. Chapter 2 Behavioral responses and ecological consequences. Fish Physiol. 2009;27:25-77.
- Sultana F, Abbas K, Xiaoyun Z, Abdullah S, Qadeer I, Hussnain RU. Microsatellite markers reveal genetic degradation in hatchery stocks of Labeo rohita. Pak J Agri Sci. 2015;52(3):775-781.
- Kabir MA, Rahman MS, Begum M, Faruque MH. Genetic diversity by rapd in four populations of Rohu labeo rohita. Croat J Fish. 2017;75(1):12-17.
- Luhariya RK, Lal KK, Singh RK, Mohindra V, Punia P, Chauhan UK, et al. Genetic divergence in wild population of Labeo rohita (Hamilton, 1822) from nine Indian rivers, analyzed through MtDNA cytochrome b region. Mol Biol Rep. 2012;39(4):3659-3665.
Citation: Tasleem F, Shahid M, Fatima H, Gulzar MN (2022) Comparative Analysis of Genetic Diversity in Natural Population of Labeo rohita and Cirrhinus marigala by Using Species Specific Molecular Markers. J Aquac Res Dev. 13:713.
Copyright: © 2022 Tasleem F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.