Indexed In
- Open J Gate
- Genamics JournalSeek
- JournalTOCs
- China National Knowledge Infrastructure (CNKI)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2023) Volume 14, Issue 6
Bioremediation Potential of Heavy Metal Multi-Tolerant Pseudomonas Spp. Isolated From a Municipal Waste Dumpsite at Ile-Ife, Osun State, Nigeria
Yetunde M Feruke-Bello1,2*, Olu Odeyemi1 and Gbolahan O Babalola12Department of Microbiology, Hallmark University, Ijebu-Itele, Ogun State, Nigeria
Received: 02-Nov-2023, Manuscript No. JPEB-23-23729; Editor assigned: 06-Nov-2023, Pre QC No. JPEB-23-23729 (PQ); Reviewed: 20-Nov-2023, QC No. JPEB-23-23729; Revised: 27-Nov-2023, Manuscript No. JPEB-23-23729 (R); Published: 04-Dec-2023, DOI: 10.4172/ 2157-7463.23.14.543
Abstract
Municipal wastes are probable source of heavy metals that may be noxious to the environment and the biotic component due to indiscriminate waste disposal method and open waste dump practiced in Africa.
Three Pseudomonas spp. isolated from a municipal waste dumpsite were evaluated for their tolerance to five metals (Pb, Cd, As, Ni and Cr). The best isolate that showed multiple resistance to all the five metals was identified as Pseudomonas aeruginosa by the 16S rRNA sequence with a Maximum Tolerable Concentrations (MTC) of 2000 mg/l for Pb, 1200 mg/l for Cd, 4500 mg/l for As, 600 mg/l for Ni and 2000 mg/l for Cr. Pseudomonas aeruginosa was subjected to mutation by ethidium bromide and acridine orange; thirteen mutant strains were recovered. The Pseudomonas aeruginosa MutAb, MutAc and MutAe exhibited a 25% increase in the MTC to lead; Pseudomonas aeruginosa MutAa-Ae recorded an improvement of 32% in the MTC to cadmium while Pseudomonas aeruginosa MutAa, MutAb, MutAd and MutAe exhibited 25% increase in the MTC to arsenic after mutational enhancement. All Pseudomonas aeruginosa strains were resistant to all the antimicrobials except for Pseudomonas aeruginosa MutAc which was sensitive to ofloxacin, nitrofurantoin and gentamicin. Pseudomonas aeruginosa MutAe exhibited the removal of 98.9% Pb from solution, compared to the Pseudomonas aeruginosa wild type with 97.4% at pH 7.
The outcome of the research specifies that the efficiency of native microbes in bio-removal procedure can be boosted by mutation for effective bioremediation of effluents with Pb metal contaminations.
Keywords
Pseudomonas aeruginosa; Bioremediation; Waste dumpsite; Enhancement; Heavy metals; Antimicrobial resistance
Introduction
Waste occurs in diverse forms such as solids and liquid; the solid forms are known as solid waste, which are produced from municipal, commercial and industrial sources [1]. According to Scarlat et al., one hundred and twenty five million tons per annum of wastes were produced in Africa in 2012 with 65% (81 million tons) of the total amount from Countries South of the Sahara, which is estimated to rise to two forty-four million tons by 2025 [2]. Wastes are frequently dumped by the public in indiscriminate places, at several dumping locations on the border of urban areas, at bare lands spread all over the city. It has been reported that haphazard and open disposal of waste can lead to environmental degradation, introduction of harmful constituents such as heavy metals into water and soil ecology, causes illnesses, unpleasant smell and threaten the wellbeing of humans and living organisms [3-5]. Regardless of the improvement in the environmental safety and ecology, the level of waste management remain inadequate [6].
Toxic metals such as arsenic (As), cadmium (Cd), mercury (Hg), and lead (Pb) have no biological role, non-biodegradable and considered a global pollution problem due to their persistence and the lethal effects in the environment and to the populace [7]. They depreciate the quality of aquatic ecosystem, poisonous at minute concentration to biological system, and causes endocrine disruption and carcinogenic [8].
There are extensive range of practices and managements to lessen or recover heavy metals from contaminated environments which include physicochemical treatment, phytoremediation and microbial bioremediation strategies [9]. Bioremediation processes have more benefits compared with conventional treatments because of the cost effectiveness and environmentally friendly. Microbes, have the competence to devour organic left-over and transform the left-over to harmless products by producing various metabolites to breakdown the difficult left-over into simple complexes [10]. Some strains of microorganisms isolated from heavy metal polluted environments have exhibited bioremediation potentials, since they have mechanisms that permit them to acclimatize to hostile environments. Some of the methods established by microbes include, metal sorption, extracellular precipitation uptake and accumulation, mineralization and enzymatic oxidation or reduction to a non-toxic form, and efflux of heavy metals from the cell [11,12].
Pseudomonas aeruginosa a Gram negative rod bacterium found in the family Pseudomonadaceae. It can adapt to the presence of some heavy metals and antibiotics [13,14]. Pseudomonas aeruginosa is one of the best bacterial strain for mitigating the effect of heavy metals pollution snags and extant in practically all polluted environment [15,16]. It has been reported as an appropriate bio-sorbent for the elimination of cadmium and other heavy metals from polluted waste, aquatic system and soil [15]. Pseudomonas aeruginosa has mastered various detoxification mechanisms, such as metal reduction and precipitation as metal salts for the elimination of heavy metals. They also produce a yellowish green fluorescence, known as Pyoverdin which are appropriate biosorbent for the elimination of cadmium and other heavy metals from polluted environment [13,15].
Despite several reports associated to the probing and bio-prospection of bacteria that may perhaps aid in bioremediation of heavy metals, there is paucity of information on the biotechnological enhancement of the native bacterial isolates from waste dump site in Nigeria for heavy metal bioremediation purposes. This study aims at isolation, characterization; identification and evaluation of tolerance mechanisms of native Pseudomonas spp. obtained from a municipal waste dump site and their mutant strains.
Materials and Methods
Bacterial strains
The Pseudomonas spp. was isolated from the soil sample of a municipal waste dump site located within a University Community at Ile-Ife. This site is a located within the co-ordinate N 7.53351, 4.52505E.
Probable identity of the multi-tolerant bacteria
Pure isolates of selected Pseudomonas spp. were identified using the Bergey’s Manual of Determinative Bacteriology [17]. Further identification was done by molecular methods. The amplification of the 16S rRNA gene sequences of Pseudomonas spp was carried out with primers 27F and 1525R, the nucleotide sequences gotten were aligned using BioEdit and related with analogous sequences at the National Center for Biotechnology Information (NCBI) with the aids of the BLAST (Basic Local Alignment Search Tool) algorithm [18].
Ethidium bromide and acridine orange stock solutions
150 mg of ethidium bromide and acridine orange were weighed and dispersed in 15 ml of deionized water, this preparation is equivalent to 10 mg/ml of the mutagen. The stock solutions were used to prepare lower concentrations of the ethidium bromide and acridine orange as required [17].
Maximum Tolerable Concentration (MTC)
The agar dilution assay was used according to the modified method of Yahaya et al. The MTC of the Pseudomonas spp. were evaluated for cadmium, lead, chromium, arsenic, and nickel. A standardized solution of the bacteria was spot inoculated to sterile Minimal Salt Agar (MSA) augmented with various concentrations of the heavy metals (200 mg/l to 5000 mg/l) after autoclaving. The control plates were set-up as positive and negative controls, all the plates were incubated at 30°C for 48 hours [19].
Generation of mutant strains
A standardized overnight culture of the Pseudomonas spp. was used for the experiment; and the methodology of Feruke-Bello et al. and Shakibaie et al [17,20]. The rate of survival of the Pseudomonas spp. Were calculated as:
%survival=Total count (cfu/ml) of mutants/total
count (cfu/ml) of wild type × 100
Antimicrobial resistance pattern of the Pseudomonas aeruginosa
The test was performed by disk diffusion assay. Eight antibiotics: ceftazidime (CAZ, 30 μg), cefuroxime (CRX, 30 μg), gentamicin (GEN, 10 μg), cefixime (CXM, 5 μg), ofloxacin (OFL, 5 μg), augmentin (AUG, 30 μg), nitrofurantoin (NIT, 300 μg), and ciprofloxacin (CPR, 5 μg) were used. A suspension of fresh culture of Pseudomonas aeruginosa was adjusted to 0.5 McFarland standards, 0.1 ml was spread on Mueller Hinton’s agar, and the antibiotic discs were placed on the surface of the plates, incubated at 37°C for 24h. The plates were examined for zone of inhibition which was measured and categorized as resistant, susceptible, or intermediate based on CLSI (2020) standards [21].
The Multiple Antibiotic Resistance (MAR) phenotypes were evaluated mathematically as:
MAR=x /y,
Where, x is the amount of antibiotics to which bacterial isolate displayed resistance and y is the sum of antibiotics which the bacterial isolate was evaluated [22,23]. The MAR index of ≥ 0.2 showed that the bacteria are from elevated risk sources while below 0.2 specified that the bacteria are from small risk sources.
The removal efficiency of cadmium by Pseudomonas aeruginosa and its mutants at different pH
The Pb removal experiment was conducted in conical flasks (250 ml) containing 50 ml MSB appended with 500 mg/l Lead at pH range of 6, 7, and 8 in an aerobic settings. A Pseudomonas aeruginosa suspension in logarithmic phase was inoculated into the solution. The solutions were agitated on orbital shaker at 170 rpm for 3-5 days. The Pb concentration in the solution after the incubation period was measured via Atomic Absorption Spectrophotometer (AAS). The %removal of lead was considered by the difference between the initial and final lead concentrations multiplied by 100 [17].
Statistical analysis
Graphpad Prism 8.4.3 was used to evaluate the statistical analysis.
Results and Discussion
Identification of the heavy metal resistant Pseudomonas spp.
The Pseudomonas spp. isolated from the municipal waste dumpsite at Ile-Ife were oxidase positive, produce pigments and fluorescence under UV light lamp. They were able to utilize various carbon sources which showed range of metabolic forms that may boost their degradability (Tables 1 and 2). Anjanapriya and Lalitha, reported the isolation of a heavy metal resistant Pseudomonas spp. from municipal solid waste dumpsites in Madurai, India [23]. Chetan et al. also reported the presence of Pseudomonas spp. from the waste dumpsite in Eagle Island [24]. The occurrence might be as a result of extensive array of physiological abilities and extracellular enzymes [17]. Which permit the organism to propagate in various environments and endure tough environmental situations in addition to polluted environments that wield selective force on the spread of particular clusters of soil bacteria. The molecular analysis of 16S rRNA gene amplification indicated that the best heavy metal resistant bacteria which showed multiple resistance to the various metals was Pseudomonas aeruginosa (GBB 215) (99% homology) when matched with related sequences in the non-redundant nucleotide databank at the NCBI with the aid of their world-wide website (BLAST).
| Bacterial code | Shape | Size | Pigmentation | Edge | Surface | Opacity | Elevation | Gram’s reaction |
|---|---|---|---|---|---|---|---|---|
| GBB 214 | Irregular | Medium | Cream | Undulate | Rough and dull | Opaque | Flat | - Rods |
| GBB 215 | Circular | Small | Cream | Entire | Smooth and glistering | Opaque | Low convex | - Rods |
| GBB 237 | Circular | Medium | Cream | Entire | Smooth and glistering | Opaque | Low convex | - Short Rods |
Table 1: Morphological features of the heavy metal resistant microbes isolated from soil of O.A.U waste dump site.
| Isolate | GBB 214 | GBB 215 | GBB 237 |
|---|---|---|---|
| Gram stain | - Rods | - Rod | - Short rod |
| Spore stain | ND | ND | ND |
| Motility test | + | + | + |
| Starch hydrolysis | - | - | - |
| Catalase test | + | + | + |
| Indole test | - | - | - |
| H2S production | + | + | + |
| Citrate utilization | + | + | + |
| Nitrate reduction | + | + | + |
| Methyl red test | + | - | - |
| Voges–Proskauer test | + | - | - |
| O/F test | O | O | O |
| Oxidase test | + | + | + |
| Glucose | A | A | A |
| Sucrose | - | - | - |
| Lactose | - | - | - |
| Maltose | - | - | - |
| Mannitol | A | A | A |
| Probable identity of Isolate | PseudomonasA sp. | PseudomonasB sp. | PseudomonasC sp. |
Table 2: The biochemical features of the heavy metal resistant microbes isolated from soil of O.A.U waste dump site.
Determination of Maximum Tolerable Concentration (MTC) of the isolated bacteria to different heavy metals in mg/l
The Maximum Tolerable Concentrations (MTC) of the Three Pseudomonas spp. were indicated in Table 3. All the isolates showed multiple resistance to all the heavy metals (Pb, As, Cd Cr and Ni), with regards to the toxicity, nickel and cadmium were the most toxic and As was the least toxic. The toxicity order was Ni>Cd>Pb>Cr>As which might be an outcome of the usage of related mechanisms for their toxicity as reported by Malik et al. [25-27].
| PseudomonasA spp. | Pseudomonas aeruginosa | ||
|---|---|---|---|
| Native isolate (removal of EtBr) | 5.40 × 106 | Native isolate (removal of acridine orange) | 1.33 × 107 |
| Mutant strains (addition of EtBr) | 4.20 × 105 | Mutant strains (addition of acridine orange) | 2.63 × 106 |
| % Survival | 7.87 | 19.77 |
Table 3: The percentage survival of the wild and mutated strains of Pseudomonas aeruginosa (GBB 215) in the presence and absence of chemical mutagen.
According to Piotrowska-Seget, et al. heavy metals may impede the development of microbes at minute concentrations and limited microbes can endure them [28]. Several investigators recounted the isolation of microbes with multiple resistance to numerous metal ions [29,30]. The results obtained in this study indicated that Pseudomonas spp. (GBB 214) had MTC of 2000 mg/l, 1200 mg/l, 4500 mg/l, 1200 mg/l and 400 mg/l for Pb, Cd, As, Cr and Ni respectively. Pseudomonas aeruginosa (GBB 215) had MTC of 2000 mg/l, 1200 mg/l, 4500 mg/l, 2000 mg/l and 600 mg/l for Pb, Cd, As, Cr and Ni respectively while Pseudomonas C spp. (GBB 237) had MTC of 1000 mg/l, 100 mg/l, 3000 mg/l, 100 mg/l and 1600 mg/l for Pb, Cd, As, Cr and Ni respectively. It was observed that isolate GBB 215 (Pseudomonas aeruginosa) had the highest tolerance to the five heavy metals with MTC of 2000 mg/l for Pb, 1200 mg/l for Cd, 4500 mg/l for As, 600 mg/l for Ni and 2000 mg/l for Cr. Several researchers have reported that Pseudomonas aeruginosa adjust to polluted environment as well tolerate heavy metals owing to its biosorption ability [31-34]. Saha and Santra, also reported the isolation of bacteria with MTC of 3000 mg/l-4000 mg/l of Pb which was higher that the MTC observed for Pb; while MTC of 10- 30 mg/l for Cd and 250-350 mg/l for As was reported; which was lower than the MTC recorded in this study [35].
The multiple and high resistance to heavy metals observed might be a reflection of the prevailing environmental condition of the soil at the waste dumpsite. Bacteria exposed to elevated amount of heavy metals develop numerous physiological and genetic mechanisms required for their adaptation and existence under such circumstances [36].
Generation of mutant
The best two bacterium that is Pseudomonas A and Pseudomonas aeruginosa were selected for further studies based on their elevated and multiple resistances to the five heavy metals. The sub-inhibitory concentration of 3.0 mg/l for ethidium bromide and 0.3 mg/l for acridine orange for Pseudomonas A and Pseudomonas aeruginosa respectively (Table 3). The colony forming unit (cfu/ ml) detected for different strains of the heavy metal resistant Pseudomonas aeruginosa are reported in Table 3. Acridine orange was identified as a superior mutagen than ethidium bromide for Pseudomonas aeruginosa [17]. Shakibaie et al. reported that acridine orange and acriflavine had extreme consequence on the bioremoval of copper and zinc [20]. Pseudomonas aeruginosa possess a survivor rate of 7.87 in ethidium bromide and 19.77 in acridine orange. A total of thirteen Pseudomonas aeruginosa mutant strains, eight from ethidium bromide and five from acridine orange were isolated from the MSA augmented with diverse concentrations of the lead salt. The generated strains of Pseudomonas aeruginosa were allotted codes based on the particular mutagen, ethidium bromide were (MutEa– MutEh) while acridine orange (MutAa-MutAe). The pigments, morphological and biochemical features of the mutant strains were not affected by the mutagen.
Evaluation of Maximum Tolerable Concentration (MTC) of the Pseudomonas aeruginosa mutant strains to different heavy metals in (mg/L)
The capability of microorganisms to endure in polluted environments has been ascribed to the structure of their cell walls which can network and bind well with heavy metals [37], in addition to a variety of genetic mechanisms that assist them to withstand the effects of the toxic metals [38].
The MTC for the heavy metal resistant bacterial isolate Pseudomonas aeruginosa Wt were 4500 mg/l, 2000 mg/l and 1200 mg/L for arsenic, lead and cadmium respectively. Feruke-Bello et al. reported a similar result in a multiple resistant heavy metal resistant Klebsiella variicola isolated from a discharged effluent in Nigeria [17]. The same MTC as that of the wild type were recorded for Pseudomonas aeruginosa MutEa–MutEh for lead while Pseudomonas aeruginosa MutAb, MutAc and MutAe recorded 25% (2500 mg/L) increase in their MTC to lead. In addition, a 25% increase (2000 mg/L) in MTC to cadmium was observed in Pseudomonas aeruginosa MutEa, Pseudomonas aeruginosa MutEd, Pseudomonas aeruginosa MutEe, Pseudomonas aeruginosa MutEf, Pseudomonas aeruginosa MutEg and Pseudomonas aeruginosa MutEh whereas 32% (2200 mg/L) increase was observed in Pseudomonas aeruginosa MutAa- Pseudomonas aeruginosa MutAe (Table 4). However, Pseudomonas aeruginosa MutEb and MutEc maintained the same MTC for cadmium as the wild type. In the presence of arsenic, the MTC observed for Pseudomonas aeruginosa MutEa-Pseudomonas aeruginosa MutEh showed an increase of 6%, while the MTC for Pseudomonas aeruginosa MutAa, Pseudomonas aeruginosa MutAb, Pseudomonas aeruginosa MutAd and Pseudomonas aeruginosa MutAe showed 25% increase.
| Mg/l | |||||
|---|---|---|---|---|---|
| Bacterial isolates | Pb | Cd | As | Ni | Cr |
| Pseudomonas aeruginosa Wt | 2000 | 1200 | 4500 | - | - |
| Pseudomonas aeruginosa MutEa | 2000 | 2000 | 4700 | - | - |
| Pseudomonas aeruginosa MutEb | 2000 | 1500 | 4700 | - | - |
| Pseudomonas aeruginosa MutEc | 2000 | 1500 | 4700 | - | - |
| Pseudomonas aeruginosa MutEd | 2000 | 2000 | 4700 | - | - |
| Pseudomonas aeruginosa MutEe | 2000 | 2000 | 4700 | - | - |
| Pseudomonas aeruginosa MutEf | 2000 | 2000 | 4700 | - | - |
| Pseudomonas aeruginosa MutEg | 2000 | 2000 | 4700 | - | - |
| Pseudomonas aeruginosa MutEh | 2000 | 2000 | 4700 | - | - |
| Pseudomonas aeruginosa MutAa | 2000 | 2200 | 4700 | - | - |
| Pseudomonas aeruginosa MutAb | 2500 | 2200 | 4700 | - | - |
| Pseudomonas aeruginosa MutAc | 2500 | 2200 | 4700 | - | - |
| Pseudomonas aeruginosa MutAd | 2000 | 2200 | 5000 | - | - |
| Pseudomonas aeruginosa MutAe | 2500 | 2200 | 5000 | - | - |
Table 4: MTC of the metal resistant wild and mutant strains of Pseudomonas aeruginosa.
Some researchers Shakibaie et al.; Feruke-Bello et al. also reported an increase in the MTC of cadmium, lead and arsenic once exposed to ethidium bromide and acridine orange by mutational boost procedures. This improvement may perhaps be as a result of the accumulation or obliteration of definite genes in the genome of the bacteria, as the chemical mutagens are alkylating agents stimulating frameshift mutation [39]. The MTC of chromium and nickel in the mutant strains were not detected after exposure to the mutagens which might be ascribed to the fact that they are plasmid encoded while lead, cadmium and arsenic resistant gene are chromosomal. Various methods existed for plasmid curing, including chemical and physical agents for the eradication of plasmid; hence ethidium bromide and acridine orange were used for the same purpose in this study.
Antibiotic sensitivity patterns of Pseudomonas aeruginosa and the mutant strains
Globally, micro-organisms resistance to antimicrobial continues to be a top menace to public health.
Municipal waste dumpsites remain a likely source of antimicrobial resistance genes due to the haphazard and incessant dumping of solid wastes in the environment [40-43].
The results for the antibiotic sensitivity profile of Pseudomonas aeruginosa and their mutant strains were presented in Table 5 according to the CLSI (2020). The Pseudomonas aeruginosa Wt (wild type) and its mutant strains showed resistance to almost all the antimicrobials used in this study except for Pseudomonas aeruginosa MutAc. The susceptibility pattern showed that 14 (100%) of Pseudomonas aeruginosa were resistant to ceftazidime, cefuroxime, cefixime and augmentin. However, 13 (92.9%) of the Pseudomonas aeruginosa and its mutant strains were resistant to gentamicin, ofloxacin and nitrofurantoin while 13 (93%) were resistant to ciprofloxacin. Oviasogie et al. also isolated a strain of P aeruginosa that was resistant to ciprofloxacin, ofloxacin, augmentin and gentamycin from municipal waste dumpsite [44].
| Pseudomonas aeruginosa strains | Antibiotics | Ceftazidime (CAZ) | Cefuroxime (CRX) | Gentamicin (GEN) | Cefixime (CXM) | Ofloxacin (OFL) | Augmentin (AUG) | Nitrofurantoin (NIT) | Ciprofloxacin (CPR) |
| 30 µg | 30 µg | 10 µg | 5 µg | 5 µg | 30 µg | 300 µig | 5µg | ||
| Wt | 0R | 0R | 4R | 0R | 4R | 0R | 3R | 14R | |
| MutEa | 0R | 0R | 4R | 0R | 4R | 0R | 0R | 14R | |
| MutEb | 0R | 0R | 4R | 0R | 5R | 0R | 0R | 15R | |
| MutEc | 0R | 0R | 4R | 0R | 5R | 0R | 0R | 15R | |
| MutEd | 0R | 0R | 3R | 0R | 4R | 0R | 0R | 15R | |
| MutEe | 0R | 0R | 4R | 0R | 5R | 0R | 0R | 14R | |
| MutEf | 0R | 0R | 4R | 0R | 5R | 0R | 0R | 14R | |
| MutEg | 0R | 0R | 5R | 0R | 5R | 0R | 0R | 15R | |
| MutEh | 0R | 0R | 3R | 0R | 4R | 0R | 0R | 14R | |
| MutAa | 0R | 0R | 4R | 0R | 6R | 0R | 4R | 15R | |
| MutAb | 0R | 0R | 5R | 0R | 4R | 0R | 4R | 15R | |
| MutAc | 0R | 0R | 19S | 0R | 20S | 0R | 21S | 21I | |
| MutAd | 0R | 0R | 4R | 0R | 0R | 0R | 0R | 17R | |
| MutAe | 0R | 0R | 4R | 0R | 0R | 0R | 0R | 13R | |
| Break Points | S | ≥ 18 | ≥ 18 | ≥ 15 | ≥ 19 | ≥ 18 | ≥ 16 | ≥ 17 | ≥ 21 |
| I | 15-17 | 15-17 | 13-14 | 16-18 | 14-17 | 13-15 | 15-16 | 16-20 | |
| R | ≤ 14 | ≤ 14 | ≤ 12 | ≤ 15 | ≤ 13 | ≤ 12 | ≤ 14 | ≤ 15 |
Table 5: The antibiotic susceptibility pattern of pseudomonas aeruginosa and their mutant strains.
In addition, 1 (7.1%) of the Pseudomonas aeruginosa which is Pseudomonas aeruginosa MutAc was sensitive to gentamicin, ofloxacin, nitrofurantoin and intermediate for ciprofloxacin (Table 5). Five classes of antibiotics (cephems, aminoglycoside, β-lactam, fluoroquinolones and nitrofurans) were used in this study. The Pseudomonas aeruginosa Wt and all the other mutants had MAR index of 1.0 except for Pseudomonas aeruginosa MutAc and Pseudomonas aeruginosa MutAd that had MAR index of 0.5 and 0.9 respectively.
A research by Hrenovic et al. established that the waste dumpsite possibly harbors the biggest and assorted resistome together with bacteria that possess intrinsic and acquired ABR [45]. Mwaikono et al. shown that municipal waste dumpsites consist of huge range and multifaceted groups of bacteria. The contact of the bacteria with heavy metals led to the choice of bacterial strain which can resist antibiotics. This occurs since genes coding for metals resistance are situated together with antimicrobial resistance genes [46]. Under situations of metal pressure, metal and antimicrobial resistance in microbes probably aids in adjusting more rapidly via range of resistant dynamics than through modification and usual selection [47].
Bioremoval efficiency of Pb by Pseudomonas aeruginosa strains
The practice of using native/wild type microorganisms in bioremediation might minimize their prospect due to antagonism and raised heavy metal concentrations. Bioremediation procedure may perhaps be enhanced by diverse methods, based on the nature of the contaminated environment [48,49]. One of such methods is the use of mutant strains of the bacteria as demonstrated by Feruke-Bello et al.; amendment of environmental variables also permit microbial development and hasten bioremediation methods [26,50].
In this study, the P. aeruginosa Wt and four of its mutants (P. aeruginosa MutAb, P. aeruginosa MutAc, P. aeruginosa MutAd, P. aeruginosa MutAe that showed higher degree of resistance to multiple heavy metal resistance were selected for further studies. The results revealed that the mutant strains P. aeruginosa MutAc and MutAe removed 96.3% and 97.1% of Pb respectively compared to 94.6% observed in the P. aeruginosa Wt at pH 6 (Figure 1). The percentage removal of Pb by P. aeruginosa MutAc and MutAe increased to 98.4% and 98.9% at pH 7 while P. aeruginosa MutAc removed 98.25% of Pb at pH 8. P. aeruginosa Wt removed 97.9% of lead from solution at pH 7 which was higher than 85% Pb removal by Pseudomonas spp. B50D in solution reported by Giovanella et al [51]. The result was also higher than 33.67% bioremediation of Pb by Pseudomonas aeruginosa as reported by Oziegbe et al [52]. It was noted that all the strains had maximum bioremediation potential at pH 7 compared with the percentage removal recorded at pH 6 and pH 8 which corroborated the report by Jin et al. that pH has a substantial outcome on the solubility of heavy metal ions and charge on the exterior of the cell which may disturbs the bio-removal procedures [53].
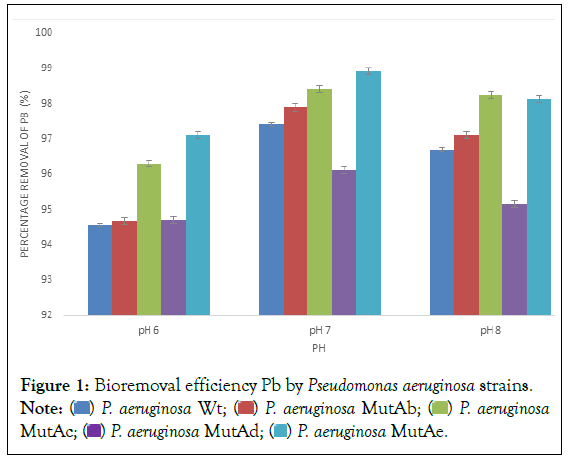
Figure 1: Bioremoval efficiency Pb by Pseudomonas aeruginosa strains. Note:  P. aeruginosa MutAc;
P. aeruginosa MutAc;  P. aeruginosa MutAe.
P. aeruginosa MutAe.
Pseudomonas spp. is reflected as one of the microbial indicator for assessing pollution in environs [41]. Pseudomonas aeruginosa has been reported to possess the capacity to repel and amass metal ions like HgCl, CuCl2 and CdCl2 [54]. Other researchers also reported that lyophilized Pseudomonas has competence for cadmium (II) and lead (II) ions uptake from solution through biosorption [55]. The mechanisms used by Pseudomonas aeruginosa in reaction to heavy metals pressure can be encrypted within the chromosomal genes, but the resistance has been frequently situated on plasmid [56].
Conclusion
Bioremediation still remains one of the utmost capable inventions for handling industrial or municipal waste comprising of heavy metals, chemical spills and hazardous wastes. The isolated Pseudomonas aeruginosa Wt showed multiple resistances to cadmium, lead, chromium, arsenic and nickel while the mutant strains Pseudomonas aeruginosa MutEa-MutEh and Pseudomonas aeruginosa MutAa- MutAe showed multiple resistances to arsenic, lead and cadmium. The resistance to chromium and nickel in the mutant strains were lost after contact with chemical mutagen (ethidium bromide and acridine orange) in a process called plasmid curing; this showed that the resistance to chromium and nickel were plasmid borne while the others are chromosomal. Although the Pseudomonas aeruginosa Wt isolated from the municipal waste dumpsite has good bioremediation potential in solution; the mutant’s strains generated from acridine orange; Pseudomonas aeruginosa MutAc and Pseudomonas aeruginosa MutAe showed a better bio-removal capability of Pb from solution, hence these strains could be a probable candidate in bioremediation and bioaccumulation of Pb from solutions.
The Pseudomonas aeruginosa strains were resistant to almost all the antibiotics in this study and the MAR index was above 0.2 which indicated that the resistance was point based pollution and from extreme antimicrobial presence in the waste dumpsite. Despite the good performance of Pseudomonas aeruginosa Wt and its mutant strains in bio-removal of Pb from solution, the probability of their usage for bioremediation may be slim due to the multiple resistances of the these strains to the commonly used antimicrobials. Antimicrobial resistance is a major menace to public health, and a main concern to global health and their presence in the environment can lead to transfer of resistant genes through the ecological structures into the food chain. The genetic alteration of native flora of a polluted environment could lead to the discovery of strains of bacteria which are good candidate for bioremediation technologies.
References
- Ojiego BO, Abdullahi SA, Gadzama IM, Bolorunduro P, Ella E, Audu KE, et al. Physicochemical and bacteriological qualities of selected solid waste dumpsites within Abuja, Nigeria.
- Scarlat N, Motola V, Dallemand JF, Monforti-Ferrario F, Mofor L. Evaluation of energy potential of municipal solid waste from African urban areas. Renew Sust Energ Rev. 2015;50:1269-1286.
- Haile T, Abiye TA. Environmental impact and vulnerability of the surface and ground water system from municipal solid waste disposal site: Koshe, Addis Ababa. Environ Earth Sci. 2012;67:71-80.
- Kebede AA, Olani DD, Edesa TG, Damtew YT. Heavy metal content and physico-chemical properties of soil around solid waste disposal sites. Amer J of sci Ind res. 2016;7(5):129-139.
- Dehghani MH, Omrani GA, Karri RR. Solid waste-sources, toxicity, and their consequences to human health. Comp Tech solid Wastewater Manag. 2021;205-213.
- Al-Khatib IA, Kontogianni S, Nabaa HA, Al-Sari MI. Public perception of hazardousness caused by current trends of municipal solid waste management. J Waste Manag. 2015;36:323-330.
[Crossref] [Google Scholar] [PubMed]
- Wasi S, Tabrez S, Ahmad M. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: A review. Environ Monit Assess. 2013;185:8147-8155.
[Crossref] [Google Scholar] [PubMed]
- Kibria G, Hossain MM, Mallick D, Lau TC, Wu R. Monitoring of metal pollution in waterways across Bangladesh and ecological and public health implications of pollution. Chemosphere. 2016;165:1-9.
[Crossref] [Google Scholar] [PubMed]
- Vidali M. Bioremediation. An overview. Pure appli chemis. 2001;73(7):1163-1172.
- Li D, Xu X, Yu H, Han X. Characterization of Pb2+ biosorption by psychrotrophic strain Pseudomonas sp. I3 isolated from permafrost soil of Mohe wetland in Northeast China. J Environ Manage. 2017;196:8-15.
[Crossref] [Google Scholar] [PubMed]
- Fu F, Wang Q. Removal of heavy metal ions from wastewaters: A review. J Environ Manage. 2011;92(3):407-418.
[Crossref] [Google Scholar] [PubMed]
- Gómez J, Gómez-Lus ML, Bas P, Ramos C, Caini F, Maestre JR, et al. ¿ Es la cuantificación del biofilm un elemento diferenciador en la patogenia de bacilos gramnegativos?. Rev Esp Quimioter. 2013;26(2).
[Crossref] [Google Scholar] [PubMed]
- K Abdul-Sada H. A resistance study of Pseudomonas aeruginosa to heavy metals. Basra J Vet Res. 2009;8(2):52-60.
- Awasthi G, Chester A, Chaturvedi R, Prakash J. Study on role of Pseudomonas aeruginosa on heavy metal bioremediation. Int J Pure App Biosci. 2015;3(4):92-100.
- Chellaiah ER. Cadmium (heavy metals) bioremediation by Pseudomonas aeruginosa: A minireview. Appl Water Sci. 2018;8(6):154.
- Abdelbary S, Elgamal MS, Farrag A. Trends in heavy metals tolerance and uptake by Pseudomonas aeruginosa. Pseudom aerug. 2019;27:1-2.
- Yetunde Mutiat FB, Gbolahan B, Olu O. A comparative study of the wild and mutated heavy metal resistant Klebsiella variicola generated for cadmium bioremediation. Bioremediat J. 2018;22(1-2):28-42.
- Altschul SF. Gapped BLAST and PSI-BLAST: A new generation of protein detabase search programs. Nucleic Acids Res. 1997;25:3389.
[Crossref] [Google Scholar] [PubMed]
- Yahaya YA, Don MM, Bhatia S. Biosorption of copper (II) onto immobilized cells of Pycnoporus sanguineus from aqueous solution: Equilibrium and kinetic studies. J Hazard Mater. 2009;161(1):189-195.
[Crossref] [Google Scholar] [PubMed]
- Shakibaie M, Khosravan A, Frahmand A, Zare S. Application of metal resistant bacteria by mutational enhancment technique for bioremediation of copper and zinc from industrial wastes. J Environ Health Sci Eng. 2008;5(4):251-256.
- Physicochemical and bacteriological qualities of selected solid waste dumpsites within Abuja, Nigeria
- Akinjogunla OJ, Enabulele IO. Virulence factors, plasmid profiling and curing analysis of multidrug resistant Staphylococcus aureus and coagulase negative Staphylococcus spp. isolated from patients with acute otitis media. Am J Sci. 2010;6(11):1022-1033.
- Anjanapriya, S., Lalitha, S. Isolation of metal resistant Bacteria from Municipal solid waste dumpsite, Madurai. Inter J of Sci Inve. 2017;6(5):659-672.
- Chetan DM, Raghavendra HL, Prithviraj HK. Isolation and Characterization of Bacteria from Solid Waste. Int j res sci innov. 2017;4(5):63-68.
- Malik A, Khan IF, Aleem A. Plasmid incidence in bacteria from agricultural and industrial soils. World J Microbiol Biotechnol. 2002;18:827-833.
- Mehrzad F, Fataei E, Rad SN, Imani AA. The investigation of nutrient addition impact on bioremediation capability of gasoil by Alcaligenes faecalis. J Pure Appl Microbiol. 2015;9(3):2185-2191.
- Mishra A, Malik A. Recent advances in microbial metal bioaccumulation. Crit Rev Environ Sci Technol. 2013;43(11):1162-1222.
[Crossref] [Google Scholar] [PubMed]
- Piotrowska-Seget Z, Cycoń M, Kozdroj J. Metal-tolerant bacteria occurring in heavily polluted soil and mine spoil. Appl Soil Ecol. 2005;28(3):237-246.
- Ndeddy Aka RJ, Babalola OO. Identification and characterization of Cr-, Cd-, and Ni-tolerant bacteria isolated from mine tailings. Bioremediat J. 2017;21(1):1-9.
- Feruke-Bello YM, Babalola G, Odeyemi O. Genetic Variability of Klebsiella Variicola by RAPD-PCR Technique and Bioremoval of Pb2+ and Cd2+ from Simulated Contaminated Soils. Intern J Soil Sedm Contam. 2022 Aug 18;31(6):770-784.
- Jia L, Zhou J, Cao J, Wu Z, Liu W, Yang C. Foam fractionation for promoting rhamnolipids production by Pseudomonas aeruginosa D1 using animal fat hydrolysate as carbon source and its application in intensifying phytoremediation. Chem Eng Process. 2020;158:108177.
- Varjani S, Upasani VN, Pandey A. Bioremediation of oily sludge polluted soil employing a novel strain of Pseudomonas aeruginosa and phytotoxicity of petroleum hydrocarbons for seed germination. Sci Total Environ. 2020;737:139766.
[Crossref] [Google Scholar] [PubMed]
- Chen Q, Li Y, Liu M, Zhu B, Mu J, Chen Z. Removal of Pb and Hg from marine intertidal sediment by using rhamnolipid biosurfactant produced by a Pseudomonas aeruginosa strain. Environ Technol Innov. 2021;22:101456.
- Al-Dhabi NA, Esmail GA, Mohammed Ghilan AK, Valan Arasu M. Optimizing the management of cadmium bioremediation capacity of metal-resistant Pseudomonas sp. strain Al-Dhabi-126 isolated from the industrial city of Saudi Arabian environment. Int J Environ Res. Public Health. 2019;16(23):4788.
[Crossref] [Google Scholar] [PubMed]
- Saha A, Santra SC. Isolation and characterization of bacteria isolated from municipal solid waste for production of industrial enzymes and waste degradation. J Microbiol Exp. 2014;1(1):1-8.
- Fashola MO, Ngole-Jeme VM, Babalola OO. Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int J Environ Res Public Health. 2016;13(11):1047.
[Crossref] [Google Scholar] [PubMed]
- Shin MN, Shim J, You Y, Myung H, Bang KS, Cho M, Kamala-Kannan S, Oh BT. Characterization of lead resistant endophytic Bacillus sp. MN3-4 and its potential for promoting lead accumulation in metal hyperaccumulator Alnus firma. J Hazard Mater. 2012;199:314-320.
[Crossref] [Google Scholar] [PubMed]
- Dupont CL, Grass G, Rensing C. Copper toxicity and the origin of bacterial resistance-new insights and applications. Metallomics. 2011;3(11):1109-1118.
[Crossref] [Google Scholar] [PubMed]
- Madigan MT, Martinko JM, Parker J. Brock biology of microorganisms.1997.
- Chen QL, Li H, Zhou XY, Zhao Y, Su JQ, Zhang X, et al. An underappreciated hotspot of antibiotic resistance: The groundwater near the municipal solid waste landfill. Sci Total Environ. 2017;609:966-973.
[Crossref] [Google Scholar] [PubMed]
- Tripathi V, Tripathi P. Antibiotic resistance genes: An emerging environmental pollutant. Persp in enviro toxi. 2017:183-201.
- Wang JY, An XL, Huang FY, Su JQ. Antibiotic resistome in a landfill leachate treatment plant and effluent-receiving river. Chemosphere. 2020;242:125207.
[Crossref] [Google Scholar] [PubMed]
- He P, Huang J, Yu Z, Xu X, Raga R, Lü F. Antibiotic resistance contamination in four Italian municipal solid waste landfills sites spanning 34 years. Chemosphere. 2021;266:129182.
[Crossref] [Google Scholar] [PubMed]
- Oviasogie FE, Ajuzie CU, Ighodaro UG. Bacterial analysis of soil from waste dumpsite. Arch Appl Sci.Res. 2010;2(5):161-167.
- Hrenovic J, Ivankovic T, Durn G, Dekic S, Kazazic S, Kisic I. Presence of carbapenem-resistant bacteria in soils affected by illegal waste dumps. Int J Environ Health Res. 2019;29(2):154-163.
[Crossref] [Google Scholar] [PubMed]
- Mwaikono KS, Maina S, Gwakisa P. Prevalence and antimicrobial resistance phenotype of enteric bacteria from a municipal dumpsite. 2015.
- Silver S, Misra TK. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42(1):717-743.
[Crossref] [Google Scholar] [PubMed]
- Ranđelović D, Stanković S, Mihailović N, Leštan D. Remediation of copper from copper mine wastes and contaminated soils using (s, s)-ethylenediaminedisuccinic acid and Acidophilic bacteria. Bioremediat J. 2015;19(3):231-238.
- Raimondo EE, Saez JM, Aparicio JD, Fuentes MS, Benimeli CS. Bioremediation of lindane-contaminated soils by combining of bioaugmentation and biostimulation: Effective scaling-up from microcosms to mesocosms. J Environ Manage. 2020;276:111309.
- Nwinyi OC, Akinmulewo BA. Remediation of soil polluted with spent oil using cow dung. Earth Environ Sci. 2019;331:012058.
- Giovanella P, Cabral L, Costa AP, de Oliveira Camargo FA, Gianello C, Bento FM. Metal resistance mechanisms in Gram-negative bacteria and their potential to remove Hg in the presence of other metals. Ecotoxicol Environ Saf. 2017;140:162-169.
[Crossref] [Google Scholar] [PubMed]
- Oziegbe O, Oluduro AO, Oziegbe EJ, Ahuekwe EF, Olorunsola SJ. Assessment of heavy metal bioremediation potential of bacterial isolates from landfill soils. Saudi J Biol Sci. 2021;28(7):3948-3956.
[Crossref] [Google Scholar] [PubMed]
- Jin Y, Luan Y, Ning Y, Wang L. Effects and mechanisms of microbial remediation of heavy metals in soil: a critical review. J Appl Sci. 2018;8(8):1336.
- Srivastava S, Agrawal SB, Mondal MK. A review on progress of heavy metal removal using adsorbents of microbial and plant origin. Environ Sci Pollut Res. 2015;22:15386-15415.
[Crossref] [Google Scholar] [PubMed]
- Kőnig-Péter A, Kocsis B, Kilár F, Pernyeszi T. Bioadsorption characteristics of Pseudomonas aeruginosa PAOI. J Serb Chem Soc. 2014;79(4):495-508.
- Abdelatey LM, Khalil WK, Ali TH, Mahrous KF. Heavy metal resistance and gene expression analysis of metal resistance genes in gram-positive and gram-negative bacteria present in Egyptian soils. J Appl Phytotechnol Environ Sanit. 2011;6(2).
Citation: Feruke-Bello YM, Odeyemi O, Babalola GO (2023) Bioremediation Potential of Heavy Metal Multi-Tolerant Pseudomonas Spp. Isolated From a Municipal Waste Dumpsite at Ile-Ife, Osun State, Nigeria. J Pet Environ Biotechnol. 14:543.
Copyright: © 2023 Feruke-Bello YM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.