Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- ResearchBible
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- MIAR
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2021) Volume 0, Issue 0
Bacterial Blood Stream Infection in Southern Ethiopia
Mengistu Hailemariam1*, Tsegaye Alemayehu1, Asnakech Agegnehu2, Mulubrhan Ali1, Enkosilassie Mitiku2 and Elshaday Azerefegne22Department of Medicine, Hawassa University Comprehensive and Specialized Hospital, Awassa, Ethiopia
Received: 22-Jan-2021 Published: 16-Feb-2021, DOI: 10.35248/2155-9597.21.s8.003
Abstract
Background: Children’s blood stream infections mainly due to resistant pathogens found to be a higher risk of hospital mortality in low and middle income countries. This study intended to assess the bacterial profile and drug resistance of isolates of blood stream infection among under- 5 children in Southern Ethiopia.
Methods: Retrospectively lab recorded data’s were retrieved to assess the bacterial isolates and rates antibiotic resistance at Hawassa University Comprehensive Specialized Hospital.
Results: Of 323 bloods culture performed 116 (35.9%) were positive for potential bacterial BSI. Positivity of blood culture decrease with age, hence neonates blood stream infection stream infection were higher than other age groups (OR, 3; 95% CI 1.5–5.1; p=0.001). The major gram positive isolate CoNS and S. aureus have shown highest level of resistance to penicillin 61.3% and tetracycline 78.8% whereas less level of resistance was reported to Meropenem 6.4%, Ceftriaxone 13% and doxycycline 13%. Although most of gram negatives isolates were resistant for tested antibiotics, K. pneumoniae has shown leaser resistance to cefotaxime and chloramphenicol.
Conclusion: A high level of blood stream infection accompanied by high drug resistant isolates was reported. The great burden in neonate than other children was reported. As a result to reduce the burden the hospital administrators need to facilities effective infection prevention and control programmes, effective hygiene practices and well-functioning environment along with educating patient about proper hygiene practices.
Keywords
Bacteria; Antimicrobial resistance; Infection
Abbrevations
ATCC: American Type Culture Collection; BSI: Blood Stream Infection; CLSI: Clinical and Laboratory Standard Institute; CoNS: Coagulase-Negative Staphylococcus; HUCSH: Hawassa University Comprehensive Specialized Hospital; IRB: Institutional Review Board; SNNPR: Southern Nation Nationalities’ and Peoples’ Region; SOP: Standard Operating Procedure; SPSS: Statistical Package for Social Sciences; WHO: World Health Organisation
Background
Blood Stream Infection (BSI) is the most common causes of morbidity and mortality in children across the world [1]. The burden of sepsis is most likely highest in low- and middle-income countries. It is estimated about 3 million newborn and 1.2 million children suffer from sepsis globally every year. Unless diagnosed on time and treated promptly, it can lead to septic shock, multiple organ failure and death. Antimicrobial resistance is a major factor determining clinical unresponsiveness to treatment and rapid evolution to sepsis and septic shock. Sepsis patients with resistant pathogens have been found to have a higher risk of hospital mortality [2].
Although, BSI can be preventable with timely attention, coherent antimicrobial therapy and supportive care, so far higher death rates reported in developing countries [3]. The rate of BSI estimated to be 3-20 times higher in developing countries due to lack of adequate and regular microbiology laboratory in these country [4].
Fever may be the only manifestation of childhood invasive infection. Nevertheless, in Ethiopia with the added burden of malaria, HIV infection, tuberculosis and malnutrition, the treatment of fever is often syndromic, targeting multiple possible aetiologies, without necessarily considering BSI as a potential cause [5]. That is why in developing country like Ethiopia, where malaria is endemic, most febrile children misleadingly managed as malarial infection and only very little proportion of them empirically managed as bacterial infection. As timely treatment of bacteremia and appropriate use of antibiotics for children is crucial most recommended replacement of presumptive treatment of malaria by test before treat as strategy for management of other febrile illness [6].
In southern Ethiopia, there is no reported data on blood stream bacterial infection among under-5 children. As a result this study provides information for local hospitals in the regions for empirical choice of antibiotics in the treatment of child bacteremia.
Methods
A retrospective cross-sectional study was conducted on all clinical sample sent to the microbiology laboratory of Hawassa University Comprehensive and Specialized Hospital (HUCSH), Hawassa, Ethiopia. HU-CSH is a teaching hospital providing both inpatient and outpatient services to more than 5 million people in the region. The microbiology laboratory is the only regional referral laboratory where patients from the region as well from parts of the region are referred for bacteriological culture and sensitivity tests. The HU-CSH microbiology laboratory was supervised and supported by the Ethiopian Public Health Institute (EPHI) quarterly. All proper sample collection for each suspected site were collected by physician or the nurses and sent to microbiology laboratory immediately. Hence, the decision to take samples for microbiological culture and the selection of samples was made by the physicians. Thus, patient related data (age and sex) with full record of bacteriological culture and antimicrobial resistance profile were retrieved from the laboratory registration book.
We retrieved all microbiological reports on bacterial pathogens from January 2019 to December 2020. The study was approved by IRB of Hawassa University College of Medicine and health sciences (HU-CMHS). Official permission from the hospital laboratory manager of Hawassa University College of Medicine and health sciences was also obtained. All laboratory methods were performed in accordance with the relevant guidelines and regulations of Clinical and Laboratory Standards Institute’s (CLSI) and accustomed to the local Standard Operation Procedures (SOPs) of our microbiology unit [7]. All data obtained in the course of the study were reserved confidential and used only for this study. We have no inclusion criteria except we reject unsuitably recorded data’s such as, illegible or incomplete age or final culture results.
Bacteriological investigation
Our laboratory bacteriological analysis follows all standard operation procedures based on the Clinical and Laboratory Standards Institute’s (CLSI). The collected clinical samples were submitted to the laboratory and processed following standard procedures. Hence according to CLSI, a 1-2 mL blood sample was collected aseptically and added to tryptic soya broth in a ratio of blood to broth 1:9 and incubated at 35°C-37°C. Visual inspection for possible microbial growth; like hemolysis, turbidity, gas production and pellicle formation was assessed until 5th days to report no bacterial growth. Turbid broth cultures were sub cultured into MacConkey agar, Blood agar plates and Chocolate agar plates (Oxoid™ Ltd, Thermo Fisher Scientific, and Waltham, MA, USA) and incubated at 37°C for 24 hours-48 hours. The Chocolate agar-incubated cultures were carried out in a microaerophilic atmosphere by using a candle jar. Bacterial identification was done primary based on colony characteristics and Gram-stain reaction followed by proper biochemical tests. Antimicrobial susceptibility profile of isolates was determined by Kirby- Bauer disc diffusion method and the results were interpreted according to CLSI guidelines [8]. The following antibiotics were used; Pencillin (10 μg), Cotrimoxazole (25 μg), Tetracycline (30 μg), Amoxicillin-Calvulanic acid (20/10 μg), Ceftazidime (30 μg), cefotaxime (30 μg), Doxycycline (30 μg), Chloramphenicol, ceftriaxone (30 μg), ampicillin (10 μg), Erythromycin (15 μg), gentamicin (10 μg), meropenem (10 μg), Amikacin (30 μg), ceftriaxone (30 μg).
Quality control
The quality of culture system was performed using E. coli (ATCC-25922), P. aeruginosa (ATCC-27853), and S. aureus (ATCC-25923) as reference strains for culture and susceptibility testing. In addition the National laboratory, Ethiopian Public Health Institute (EPHI) supervised as well supplies all necessary quality issues.
Data analysis
The data was entered and analysed using SPSS version 20 (IBM Corporation, Armonk, NY, USA). Descriptive statistics like frequency and percentages of categorical variables was calculated. Bivariate comparisons using Chi-square logistic regression were employed to assess the association between variables. A p-value of less than 0.05 was considered as statistical significance.
Results
We processed a total of 323 blood cultures from child who were suspected for blood stream infection during the study period. The ratio of male were 172 (53.3%) and 151 (46.7%) females. From all processed clinical samples, 116/323 (35.9%) were culture positive for invasive bacterial infection. Among recorded 116 bacterial infections, 62 (53.4%) were male and 54 (46.6%) female. Dominant number of patients, 142 (44%) as well the highest positivity rate, 52.6% was recorded among the age group of 0-28 day neonates, p=0.003
Bacterial isolates
In this study a total of 116 bacteria were documented of these 62 (53.4%) were gram positive bacteria. The frequent isolate were; CoNS 29 (24.8%), Klebsiella pneumoniae 25 (21.4%), and S. aureus 21 (17.9%) while proteus and entrobacter with 2 (1.7%) were the least isolate found (Figure 1). From the isolated, Grampositive bacteria were more prevalent than Gram-negative bacteria
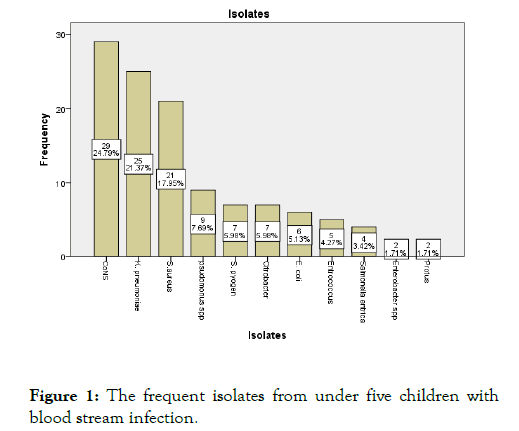
Figure 1: The frequent isolates from under five children with blood stream infection.
Antibiotic resistance of gram positive
As antibiotic resistance summaries of gram positive bacterial isolates shown, most of the isolates recorded resistance greater than 60% for penicillin, tetracycline and Erythromycin. S. aureus and CoNS show the list resistance to ceftriaxone, cefotaxime and Meropenem while commonly used drugs shows highest resistance. S. aureus also show highest resistances to, Penicillin, Tetracycline and Erythromycin. Similarly Enterococci reported list resistance to ceftriaxone, meropenem and doxycycline.
Antibiotic resistance pattern of gram negatives
Yet again the most frequent isolate, K. pneumoniae recorded the high resistance almost for all common antibiotics mainly to ampicillin, ceftazidime and ciprofloxacin however, least resistance documented to chloramphenicol and cefotaxime. In contrast P. aeruginosa and E. coli confirmed highest resistance to chloramphenicol although still they showed high resistance for the rest of other antibiotics. Ceftriaxone seems the least resisted drug for E. coli and cefotaxime and ceftriaxone appears least resisted by other isolates of gram negatives .
Discussion
Bacterial blood stream infection of children is one of the most serious problems worldwide [2]. The lack of proper microbiological analysis in developing country like Ethiopia has challenged the process of reducing the burden. This study revealed that a highest prevalence of (35.9%) bacterial BSI among children compared to similar studies in different corner of the country; Jimma 8.8%, Addis Ababa 32.8% and other African countries like Tanzania 14.2%; 24%, Gabon 17%, Burkina Faso 6% in S. Africa 5.5%; 5.7% and among Iranian neonates 6.4% [9-16,5]. Our results are comparable to study done in Gondar, northern Ethiopia (39.5.9%) [17]. While it was lower compared to the study done in Gondar 46.6% [18] among neonates suspected to sepsis. Possible reason for variation might be the variation in clinical indication for blood culture and patient age group. In our set up blood culture demanded after explicate diagnosis of other doubts due to the fact that blood culture is costly and take long time. Moreover poor effective hygiene practices and well-functioning environment also poor patient hygienic practices might increase the rate of infection.
In Ethiopia, bacterial sepsis was reported as the leading cause of neonatal death [19]. One of the systematic review studies in Ethiopia has reported that a pooled prevalence of neonatal sepsis in Ethiopia was 49.98% [20]. Likewise in this study higher rate of blood stream infection recorded with the odds of neonates being more positive in blood stream infection than other age group of under 5 children (OR, 3; 95% CI 1.5-5.1; p=0.001). Most other study also reported neonates BSI higher compared to [17,20].
Regarding isolate, the current study has found that almost 43% of the isolates were CoNS and S. aureus followed by Klebsiella 21.4% which is similar with other studies in Ethiopia, in Arsi and Gondar also with most of other studies in Africa [17,21]. When we look report from African study, BSI study on neonatal in Tanzania reported that S. aureus 36.5% and Klebsiella 29.7% dominate all isolates recognized there is also agreement with report from S. Africa were S. aureus, Klebsiella and E. coli are predominant isolate BSI among children [5]. Type of isolates in this study is still similar to that reported in other African studies for instant report from S. Africa shows predominate isolates S.aureus, Klebsiella among blood cultured samples of children. Again in Zimbabwe, CoNS and S.aureus were dominant [15,22]. This might show sort of nosocomial infection due to procedural insertion of different medical device to child. At the mean time health workers poor effective hygiene practices and wellfunctioning environment that can source birth canal contamination of Klebsiella to neonatal [22]. likewise to this study a systematic review study done in resource limited countries had also reported that Gram-negative organisms, Klebsiella pneumoniae was dominant isolate from child blood culture. In the other hand study from Ghana reported nontyphoidal salmonellae was dominant [23,24].
As usual gram positive isolates shows highest level of resistance to penicillin 61.3% and tetracycline 78.8% which is similar with report in Jimma and Gondar also comparable with report of systematic review which covers most of the regions found in Ethiopia [9,17,25]. S aureus and CoNS shown high level of resistance penicillin (59%, 71.4%), Amoxicillin and Calvulanic acid (20.6%, 19%), tetracycline (80%) and Erythromycin (62%, 71.4%), which is again similar with systematic review done in Ethiopia [26]. However less level of resistance was reported to Meropenem (6.4), Ceftriaxone (13%) and doxycycline (13%) that is comparable with previous study in the region [27].
The overall BSI rates of gram negative isolates were 17% and almost all resisted Ampicillin (80%), Ciprofloxacin (61.2%) and Ceftazidime (65.5%). As displayed in 4, most of the isolates are resistant for the tested antibiotics 36%-80% of resistance. The predominate isolate, K. pneumoniae have shown leaser resistance tocefotaxime and chloramphenicol while P. aeruginosa and E.coli resist more than 50%. Though comparison of our finding with others report is difficult due to the fact that antibiotic resistance vary place to place in addition to lack of consistency in the measurement and reporting of susceptibility data makes it difficult to compare findings among different countries and laboratories, sometimes even within one country [28]. But most of the study revealed high resistance of gram negatives. In fact in resource limited countries large regional variations in resistance rates to locally prescribed antibiotics is common [17,23]. Due to concerning other gram negative isolates we are unable to discuss on rate of resistance due to leaser number of observation.
Conclusion
In general the finding in this study indicates high level of BSI among under-five children. A high drug resistance to locally used antibiotics also surge the challenge. Therefore we recommend for local health professionals to see choices of best drug from reported compiled date for their routine empirical treatment of blood stream bacterial infections. Furthermore the hospital administrators should facilities mainly on having functioning infection prevention and control programmes, effective hygiene practices and precautions, along with a clean, well-functioning environment and equipment also educating patient about effective hygiene practices.
Ethics approval and consent to participate
This study was approved by IRB of Hawassa University College of Medicine and health sciences. Administrative permission to access data was requested through the hospital laboratory manager of Hawassa University College of Medicine and health sciences. We have got waiver from hospital laboratory and all data obtained in the course of the study were reserved confidential and used only for this study.
Limitations
First as it is retrospective study from registered in laboratory log book we didn’t have different documented independent variables for possible causal effect association. Hence we did not investigate risk factor for infection as well for drug resistance. Second, due to lack of appropriate facility anaerobic were not included. Third we have excluded vancomycin resistance test due to lack of uniformity.
Acknowledgements
We would wish to express our genuine thankfulness to the staff of microbiology lab for reliable help during compiling of data from log books. We also thank Hawassa university comprehensive specialized hospital for official permission of this study.
Authors’ Contributions
All authors; MH, TA, EA, AA, MA, and EM: participated in conception of the study, collected the data, interpretation of data and drafting the manuscript. MH and TA: design of study and critically reviewing the manuscript for important intellectual content. All authors read and approved the final manuscript for publication.
Funding
Not applicable
Competing of interests
The authors declare that they have no competing interests.
Availability of Data and Materials
The data that support the findings of this study are available from the corresponding author on upon reasonable request
Consent for Publication
Not applicable.
REFERENCES
- Liu L, Oza S, Hogan D, Chu Y, Perin J, Zhu J, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the sustainable development goals. Lancet. 2016;388(10063):3027-3035.
- Fleischmann-Struzek C, Goldfarb DM, Schlattmann P, Schlapbach LJ, Reinhart K, Kissoon N. The global burden of paediatric and neonatal sepsis: a systematic review. Lancet Respir Med. 2018;6(3):223-230.
- Labi A-K, Obeng-Nkrumah N, Bjerrum S, Enweronu-Laryea C, Newman MJ. Neonatal bloodstream infections in a Ghanaian Tertiary Hospital: Are the current antibiotic recommendations adequate?. BMC Infect Dis. 2016;16(1):598.
- Reiner RC, Olsen HE, Ikeda CT, Echko MM, Ballestreros KE, Manguerra H, et al. Diseases, injuries, and risk factors in child and adolescent health, 1990 to 2017 findings from the global burden of diseases, injuries, and risk factors 2017 study. JAMA Pediatr. 2019;173(6):e190337.
- Lochan H, Pillay V, Bamford C, Nuttall J, Eley B. Bloodstream infections at a tertiary level paediatric hospital in South Africa. BMC Infect Dis. 2017;17(1):750.
- Muro F, Reyburn R, Reyburn H. Acute respiratory infection and bacteraemia as causes of non-malarial febrile illness in African children: a narrative review. Pneumonia (Nathan). 2015;6(1):6-17.
- Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute. 2017.
- Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement. Clinical and Laboratory Standards Institute. 2018.
- Zenebe T, Kannan S, Yilma D, Beyene G. Invasive bacterial pathogens and their antibiotic susceptibility patterns in Jimma university specialized hospital, Jimma, Southwest Ethiopia. Ethiop J Health Sci. 2011;21(1):1-8.
- Kitila KT, Taddese BD, Hailu TK, Sori LM, Geleto SE, Mengistu GZ, et al. Assessment of bacterial profile and antimicrobial resistance pattern of bacterial isolates from blood culture in Addis Ababa regional laboratory, Addis Ababa, Ethiopia. Clin Microbiol. 2018;7(2):312.
- Seni J, Mwakyoma AA, Mashuda F, Marando R, Ahmed M, DeVinney R, et al. Deciphering risk factors for blood stream infections, bacteria species and antimicrobial resistance profiles among children under five years of age in North-Western Tanzania: a multicentre study in a cascade of referral health care system. BMC Pediatrics. 2019;19(2):32.
- Mhada TV, Fredrick F, Matee MI, Massawe A. Neonatal sepsis at Muhimbili national hospital, Dar es Salaam, Tanzania; aetiology, antimicrobial sensitivity pattern and clinical outcome. BMC Public Health. 2012;12(5):904.
- Alabi AS, Frielinghaus L, Kaba H, Kösters K, Huson MAM, Kahl BC, et al. Retrospective analysis of antimicrobial resistance and bacterial spectrum of infection in Gabon, Central Africa. BMC Infect Dis. 2013;13(1):455.
- Kiemde F, Tahita MC, Lompo P, Rouamba T, Some AM, Tinto H, et al. Treatable causes of fever among children under five years in a seasonal malaria transmission area in Burkina Faso. nfect Dis Poverty. 2018;7(1):60.
- Dramowski A, Cotton MF, Rabie H, Whitelaw A. Trends in paediatric bloodstream infections at a South African referral hospital. BMC Pediatr. 2015;15(1):33.
- Nikkhoo B, Lahurpur F, Delpisheh A, Rasouli MA, Afkhamzadeh A. Neonatal blood stream infections in tertiary referral hospitals in Kurdistan, Iran. Ital J Pediatr. 2015;41(1):43.
- Abebaw A, Tesera H, Belachew T, Mihiretie GD. The bacterial profile and antibiotic susceptibility pattern among patients with suspected bloodstream infections, Gondar, north-west Ethiopia. Pathol Lab Med Int. 2018;10(1):1-7.
- G/eyesus T, Moges F, Eshetie S, Yeshitela B, Abate E. Bacterial etiologic agents causing neonatal sepsis and associated risk factors in Gondar, Northwest Ethiopia. BMC Pediatr. 2017;17(1):137.
- Assefa N, Lakew Y, Belay B, Kedir H, Zelalem D, Baraki N, et al. Neonatal mortality and causes of death in Kersa health and demographic surveillance system (Kersa HDSS), Ethiopia, 2008–2013. Matern Health Neonatol Perinatol. 2016;2(1):7.
- Belachew A, Tewabe T. Neonatal sepsis and its association with birth weight and gestational age among admitted neonates in Ethiopia: systematic review and meta-analysis. BMC Pediatrics. 2020;20(1):55.
- Sorsa A. Epidemiology of neonatal sepsis and associated factors implicated: observational study at neonatal intensive care unit of Arsi university teaching and referral hospital, South East Ethiopia. Ethiop J Health Sci. 2019;29(3):333-342.
- Nathoo KJ, Chigonde S, Nhembe M, Ali MH, Mason PR. Community-acquired bacteremia in human immunodeficiency virus-infected children in Harare, Zimbabwe. Pediatr Infect Dis J. 1996;15(12):1092-1097.
- Le Doare K, Bielicki J, Heath PT, Sharland M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Soc. 2015;4(1):11-20.
- Nielsen MV, Sarpong N, Krumkamp R, Dekker D, Loag W, Amemasor S, et al. Incidence and characteristics of bacteremia among children in rural Ghana. PLoS One. 2012;7(9):e44063.
- Reta A, Kifilie AB, Mengist A. Bacterial infections and their antibiotic resistance pattern in Ethiopia: A systematic review. Adv Prev Med. 2019;2019(1):4380309.
- Deyno S, Fekadu S, Seyfe S. Prevalence and antimicrobial resistance of coagulase negative staphylococci clinical isolates from Ethiopia: a meta-analysis. BMC Microbiol. 2018;18(1):43.
- Tsegay E, Hailesilassie A, Hailekiros H, Niguse S, Saravanan M, Abdulkader M. Bacterial isolates and drug susceptibility pattern of sterile body fluids from tertiary hospital, Northern Ethiopia: A four-year retrospective study. J Pathog. 2019;2019(1):5456067.
- Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, González IJ, et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17(1):616.
Citation: Hailemariam M, Alemayehu T, Agegnehu A, Ali M, Mitiku E, Azerefegne E (2021) Bacterial Blood Stream Infection in Southern Ethiopia. J Bacteriol Parasitol. S8: 003.
Copyright: © 2021 Hailemariam M , et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

