Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- JournalTOCs
- CiteFactor
- Ulrich's Periodicals Directory
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- Centre for Agriculture and Biosciences International (CABI)
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2020) Volume 11, Issue 3
Application and Evaluation of the Loop-Mediated Isothermal Amplification assay for the Detection of Aflatoxigenic Fungi Contaminants of Rice Grains in Kenya
Youmma Douksouna1*, Allan Ole Kwallah2, Andrew Nyerere3, Steven Runo4 and Zachée Ambang52Centre for Clinical Research, Kenya Medical Research Institute, Nairobi, Kenya
3Department of Medical Microbiology, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya
4Department of Biochemistry, Microbiology and Biotechnology, Kenyatta University, Nairobi, Kenya
5Department of Plant Biology and Biotechnology, University of Yaounde, Cameroon
Received: 28-Jan-2020 Published: 20-Mar-2020, DOI: 10.35248/2157-7471.20.11.491
Abstract
Aflatoxigenic fungi are most common filamentous fungi that synthesis aflatoxins and represent the major fungal pathogens to agricultural products. Aflatoxins remain a major threat to global food security, these molecules could be resisted into food during processing and in additional may remain within the food chain. Aflatoxins are carcinogenic, hepatotoxic, mutagenic, teratogenic, can inhibit numerous metabolic systems and immunosuppressive properties. Studies of aflatoxigenic strains can help to enhance strategies control and prevent aflatoxigenic fungi contamination and aflatoxins production in foodstuffs. In this study, isolation of Aspergillus species was based on morphological characteristics including the mycelium growth pattern, color, and properties of fruiting bodies of the fungi. The innovated technique loop-mediated isothermal amplification assay was applied to amplify Norsolorinic Acid gene. The loop-mediated isothermal amplification have been optimized by combination of the rapidity, simplicity and specificity to detect the target genomic DNA in the reactions. The amplification curves monitored by Loopamp realtime Turbidimeter were analyzed in order to distinguish aflatoxigenic and non-aflatoxigenic strains.
Overall, the results showed that the loop-mediated isothermal amplification method was effective in detecting aflatoxigenic strains with high specificity of 71.5% and sensitivity under lower concentrations of DNA. In additional, it was faster than the conventional polymerase chain reaction. The loop-mediated isothermal amplification assay described in this study might be a promising tool for prediction potential threats by aflatoxigenic fungi and aflatoxins risk in food and commodities.
Keywords
Rice grains; LAMP; Rapid detection; Nor-1; Aflatoxigenic
Introduction
Aflatoxigenic fungi is one of the greatest amount the contaminants of food and commodities including cereals [1]. Rice grains, like other cereals, can be noticed as an appropriate substrate for fungal growth [2]. Under warm and dry climatic conditions, rice grains as raw material, can be invaded by aflatoxigenic Aspergillus species (spp.) [3]. Aspergillus belongs to phylum Ascomycota, kingdom of Fungi. They are ubiquitous and universal in ecosystem, found as saprophytes in decaying vegetation, grains seed [4]. In the environment, Aspergillus exists as conidia or sclerotia and in plant tissues as mycelia, can contaminate food crops with severely secondary metabolites [5]. Numerous species in Aspergillus section flavi are frequently commonly in plants and their processed derivatives, with some producing diverse metabolites, such as aflatoxins, 3- nitropropionic acid, tenuazonic acid, and cyclopiazonic acid [6]. Aflatoxins (AFs) productions are associated with spore production by species of Aspergillus (aflatoxigenic strains) [7].
Rice (Oryza sativa L.) is mostly consumed in Sub Saharan Africa and is one of the important foods. Several investigations reported the prevalence of mycotoxigenic molds and mycotoxin occurrence in rice; in particularly Aspergillus spp. [8,9] and their possible related metabolites have been investigated [10]. The maximum limits for these mycotoxins in rice were set by the European Commission [11]. Several data about appearance of mycotoxin contamination problems of some countries in Africa were been reported by international research institutions particularly the International Agency for Cancer Research (IARC) [12]. AFs are carcinogenic, hepatotoxic, mutagenic, teratogenic, immunosuppression properties and can inhibit many metabolic systems.
About 5.2 million cancer deaths occur each year, 55% of which arise in developing countries. In sub Saharan Africa, 25,000 hepatocellular carcinoma related deaths appear frequently because of aflatoxins ingestion [13]. The dismissal of AFs is difficult because of their stability molecules and thermal resistive in dried products [14]. Additionally, AFs are resistant throughout the foodstuffs processing and could be remained along the food chain [15]. Therefore, AFs consider a potential challenge and threat to human health, both by the direct ingestion of contaminated food products or by carry over aflatoxins and their substances in milk and meat [16]. World health authorities warn that low doses with long-term dietary exposure to aflatoxins represent a major risk of hepatocellular carcinoma [17].
The presence of any species of Aspergillus section flavi mentioned above into food and commodities may indicate aflatoxigenic contaminants. Therefore, detection of aflatoxigenic strains and the estimation of the levels of aflatoxin in infected rice grains is important as this will mean indicate food quality as well as of the potential future risk of aflatoxins present in food [18]. To achieve this, molecular tools such as PCR based techniques were developed and applied for the detection of pathogenic fungi by amplifying their housekeeping genes [19]. These include genes coding for ribosomal RNA [20] or, most commonly, those that are involved in the aflatoxins biosynthesis pathways [21]. Recently, DNA based biosensors were developed to detect aflatoxigenic fungi [22]. Nevertheless, these techniques of detection are costly, time consuming, labor intensive requires personnel training and special equipment. Additionally, reagents hamper the broad practical application of PCR based techniques in low tech environment and in the field. Another hinder is the fact that PCR based techniques need regularly some quality of DNA clean up and concentration prior for analysis. Moreover, directly detection of contaminants from colonized commodities by PCR is mostly hampered by the complex mycobiota frequently containing closely related species and a complex mixture of compounds that could affect the efficiency and sensitivity of a PCR assays [23].
As an alternative, a new specific, rapid, cost-effective and easy to use technique called loop-mediated isothermal amplification (LAMP) method of DNA was developed [24]. Ludwig et al. [25] and Jie et al. [26] reported the application of the new technique for the detection of aflatoxigenic fungi in food quality control respectively. LAMP makes use of 4/6 primers that recognize 6/8 different binding regions in the target DNA which are required to bind properly in order to initiate DNA amplification by a thermophilic DNA polymerase with high strand displacement activity and provides very high specificity [24,27]. It based upon auto cycling strand displacement DNA synthesis with the existence of Bst polymerase in isothermal circumstances for 60 min at constancy temperature [24].
Therefore, the aim of this study was to apply and evaluate the LAMP assay for detection aflatoxigenic strains of Aspergillus species contaminants rice grains. The adopted protocol offers a rapid, high specificity and sensitivity for the detection approach of aflatoxigenic fungi and has a great potential to simplify mycotoxigenic species monitoring and mitigating in food raw materials.
Materials and Methods
Samples
Samples comprising local and imported rice grains were collected randomly (1 kg) from local retail markets and millers in Mwea and Thika (Kenya) and labelled appropriately. A total of 98 samples (local rice produced in Mwea and imported rice originating from Biriyani, India, Pakistan, and Thailand) were taken according to the alternative sampling plan for the official control of mycotoxins in food [11]. Representative samples were then put in sealed bags and transported to the Molecular Biology and Biotechnology Laboratory in Pan African University institute for Basic Sciences, Technology and Innovation (PAUSTI).
Fungal cultures
Samples of rice grains were analyzed by the direct plating method described by Pitt et al. [28]. Isolation of fungi from the suspected grains was conducted following described method of Ulster [29]. Isolation of emerging fungi colonies in modified rose Bengal chloramphenicol agar (MRBA) was done following [30] method. Pure cultures were done according to Pitt et al. [30] and Varga et al. [31] for subsequent studies on two (2) culture media: malt extract agar (MEA) and potato dextrose agar (PDA), for seven days followed by phenotypic Characterization of Aspergillus species.
DNA extraction
The genomic DNA was extracted from seven-day-old fungi cultures grown on MEA and PDA media and the fungal mass from the pure cultures was scraped-out from the plates. A total of 50–100 mg of fungal mycelium was scraped and placed in a 2 mL tube and vortexed vigorously for 30 min with glass beads to crush the mycelial wall and to release the DNA. A volume of 500 μL of Lysis Buffer (100 mM Tris-HCl pH 8, 1 mM EDTA, 100 mM NaCl, 10 mM B-mercaptoethanol, and 1% Sodium dodecyl sulfate), 5 μL of RNase A, and 1 μL of Proteinase K were added. Tubes were then incubated at 65°C for 45 min in buffer. Thereafter, 270 μL of Sodium/Potassium acetate (3 M) was added and samples were well-mixed and centrifuged at 13,000 rpm for 10 min. After centrifugation, the supernatant (700 μL) was transferred to a fresh tube, and an equal volume of chloroform: isoamylalcohol (24:1) was added and mixed well, samples were stood on a bench for 5 min, followed by centrifugation at 13,000 rpm for 10 min. The supernatant (700 μL) was transferred into new tubes, and 80 μL of Sodium/ Potassium acetate (3 M) and 587 μL of ice cold isopropanol were added and then mixed well by inverting the tubes and then incubating them at -20°C overnight. After that, samples were centrifuged at 13,000 rpm for 30 min, and the supernatant was discarded carefully. The DNA pellets were washed with 1 mL of 70% ethanol and centrifuged at 13,000 rpm for 10 min. The DNA pellets were air dried and dissolved in 50 μL of Tris-EDTA (TE) buffer and stored at -20°C until use. The concentration and purity of genomic DNA were measured using a Nano-drop Spectrophotometer (Model PCR Max Lambda, Thermo Fisher Scientific, Waltham, MA, USA), and the quality of all extracted DNA was measured by taking their absorbance at 260 nm and 280 nm.
LAMP primers used in this study to detect aflatoxigenic strains
Specific primer sequences reported/designed by Ludwig et al., [25] were used to amplify the gene of interest involved in the aflatoxin biosynthesis pathway Norsolorinic Acid (NOR): aflD (nor-1), fragment of aflatoxigenic fungal genomic DNA. The primers were synthesized and purified by Macrogen Korea 10 F, 254 Beotkkot-ro Geumcheon-gu, Seoul 08511, Republic of Korea. The sequences of primers are listed in Table 1.
| Primer name | Sequences (5'–3') |
|---|---|
| F3 | ACT GCG ACT CGG AAA GYG A |
| B3 | GGA CTG CTG CAG CAT CAG |
| FIP | GGC CCA AAG TTC TGC GCC ATC CAG ACA TTG CGG GAR GA |
| BIP | ACC ATG CCC CTC GAR CAT CTG CGG GTT GCC TGA AAC AG |
| LF | ACY ACC ACG TCC AAG TGC |
| LB | TGA TGG TCA AYA TGT ATG CTC C |
Table 1: Oligonucleotide primer sets used for the study.
PCR detection
PCR analysis was performed in a 20 μL reaction mixture comprising 1 μL of genomic DNA, 4 μL of premix Taq buffer (Solis Biodyne, Tartu, Estonia), and 0.5 μL of each the forward and reverse primer (F3 and B3). The final volume was topped up to 20 μL with nuclease free water. Amplification was performed in a Proflex PCR System (Model 4483636, Thermo Fisher Scientific, Waltham, MA, USA) with the following cycling conditions: an initial denaturation at 95°C for 5 min, followed by 33 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 45 s, Elongation at 72°C for 45 s and final Elongation 72°C for 7 min. The amplicons were resolved by gel electrophoresis in a 1.5% agarose (Sigma, St. Loaus, MO, USA) gel stained with 0.1 μL/ml of Trugel Fluorescent Dye. The DNA bands resolved on agarose gel were visualized in the UV-gel documentation system (Model UV Doc. HDS UITEC Cambridge, UK). The sizes of the amplicon were estimated by comparing them with a commercial Cleaver CLS-MDNA-1 kb DNA ladder RTU 1151021805 on agarose gel.
Analytical specificity and sensitivity of LAMP assay
RNA Amplification kit (RT-LAMP) was obtained from Eiken Chemical Co., Japan to test specificity and sensitivity of the LAMP assay following the manufacturer’s instructions. Briefly, 25 μL containing reaction comprising 12.5 μL of Reaction Mix (RM), 7.7 μL of Distilled Water (DW), 0.4 μL each FIP and BIP, 0.2 μL each LF and LB, 0.05 μL each F3 and B3 primers, 1.0 μL of Enzyme Mix (EM) and 2.5 μL of test sample (extracted DNA) was set-up. Reactions were incubated at 61°C for 1 hour in a heat Blocks (A&B) using Loopamp LA-500 real-time Turbidimeter (SNEAOZO259, EIKEN Chemical Co., LTD, Tokyo, Japan) after optimizing conditions. To evaluate the analytical sensitivity (limit of detection) of the LAMP assay, positive samples in specific test were diluted to obtain 7-point, 10-fold dilutions. Loopamp LA-500 real-time Turbidimeter and gel electrophoresis were used to examine the reaction results. The LAMP amplicons were analyzed by gel electrophoresis in a 1.5% agarose (Sigma, St. Loaus, MO, USA) gel stained with 0.1 μL/ml Trugel Fluorescent Dye in a 100 ml of TAE buffer. The DNA bands resolved on agarose gel were visualized in the UV gel-documentation system (Model UV Doc. HDS UITEC Cambridge, UK). For electrophoresis analysis, 1 kb Plus DNA Ladder (Thermo Scientific Gene Ruler DNA Ladder Col 160980418) was used as a DNA size marker. The LAMP results were then drawn to judge whether the reaction was positive or negative and presented as amplification curves in graphs generated by GraphPad Prism Software version 7.00.
Results
Isolation of Aspergillus species
Based on the phenotypic characteristics including the mycelium growth pattern, color, and properties of fruiting bodies of the fungi, a total of 258 Aspergillus were isolated from rice grains in this study. Among the predominant species identified are Aspergillus clavatus, Aspergillus flavus, Aspergillus fumigatus, Aspergillus nomius, Aspergillus oryzae, Aspergillus parasiticus, Aspergillus versicolor and Aspergillus spp. These strains were purified according on their distinct morphology on MEA and PDA plates and differentiated based on their macroscopic and microscopic characteristics. Macroscopic identification was based on colony and reverse color, diameter, exudates, and texture. On the basis of the macroscopic study, isolates could not be differentiated. Further microscopic study involved the arrangement, color, diameter, shape, size, wall characters, cellular contents, conidial heads, conidiophore, sterigmata, conidia, and conidial arrangements, which were observed with a microscope. All these characteristics indicated that they were the species of Aspergillus. The macroscopy and microscopy characteristics of Aspergillus isolates are listed in Figure 1.
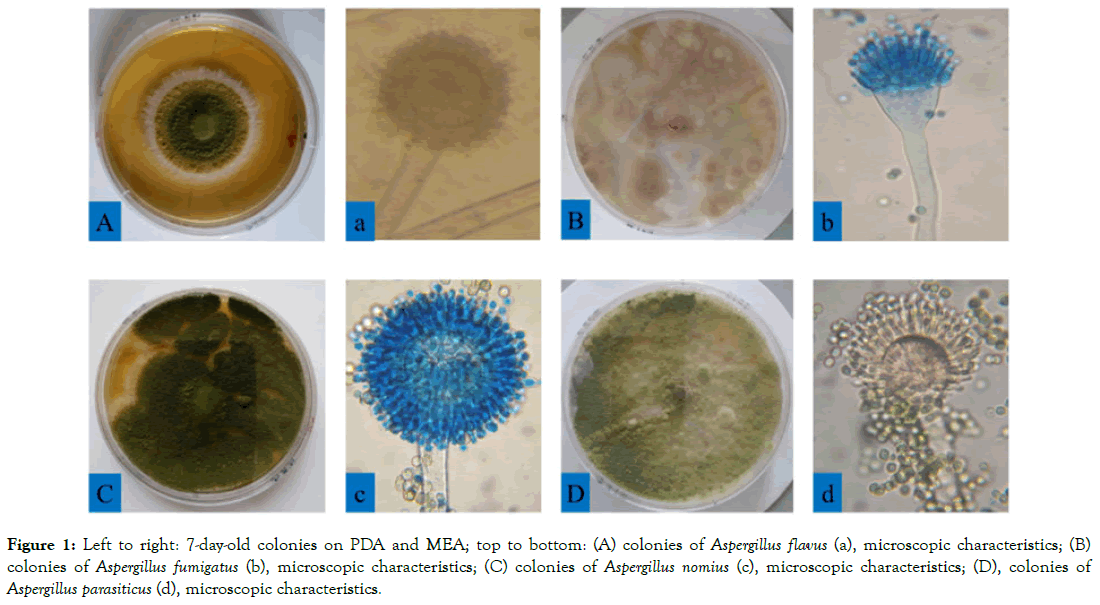
Figure 1: Left to right: 7-day-old colonies on PDA and MEA; top to bottom: (A) colonies of Aspergillus flavus (a), microscopic characteristics; (B) colonies of Aspergillus fumigatus (b), microscopic characteristics; (C) colonies of Aspergillus nomius (c), microscopic characteristics; (D), colonies of Aspergillus parasiticus (d), microscopic characteristics.
Based on morphological characteristics, analysis revealed that Aspergillus flavus was the most represented strain, followed by Aspergillus parasiticus, Aspergillus fumigatus, Aspergillus nomius Aspergillus oryzae, Aspergillus clavatus, Aspergillus versicolor and Aspergillus spp. as shown in Table 2.
| Strains | Isolates | Percentage (%) |
|---|---|---|
| Aspergillus clavatus | 08 | 3.1 |
| Aspergillus flavus | 97 | 37.7 |
| Aspergillus fumigatus | 28 | 10.8 |
| Aspergillus nomius | 16 | 6.2 |
| Aspergillus parasiticus | 72 | 27.9 |
| Aspergillus oryzae | 13 | 5 |
| Aspergillus versicolor | 07 | 2.7 |
| Aspergillus spp | 17 | 6.5 |
Table 2: Aspergillus species isolated after pure culture.
Aflatoxigenic screening
A total of 137 genomic DNA was isolated from pure cultures of Aspergillus species isolated from r ice grains for aflatoxigenic analysis. The quantity (280/260 ratio) of extracted DNA from all the samples was between 369 μg/mL and 1998 μg/mL, which indicating a good quality material for downstream processes. To evaluate the specificity and sensitivity of PCR and LAMP assays, 137 samples have been screened to detect aflatoxigenic strains by amplification the Norsolorinic Acid (NOR): aflD (nor-1) gene, involved in the aflatoxin biosynthetic pathway.
Inspection of the LAMP assay product
In order to determine the applicability of the LAMP in detection of aflatoxigenic fungi, 7 isolates were used in the reaction for optimization of the LAMP technique conditions and one reaction without template (DNA) served as negative control. The reaction curves indicated that the primers set were able to amplify the target DNA sequence of aflatoxigenic species. The preliminary experiment showed 5 positive amplification results of the LAMP amplification through records of absorbance at 400 nm under a Loopamp LA-500 real-time Turbidimeter (SNEAOZO259, EIKEN Chemical Co., LTD, Tokyo, Japan), in contrast, the negative reaction samples remained parallel to the abscissa axis as shown by Loopamp LA-500 real-time Turbidimeter analysis of curves (Figure 2).
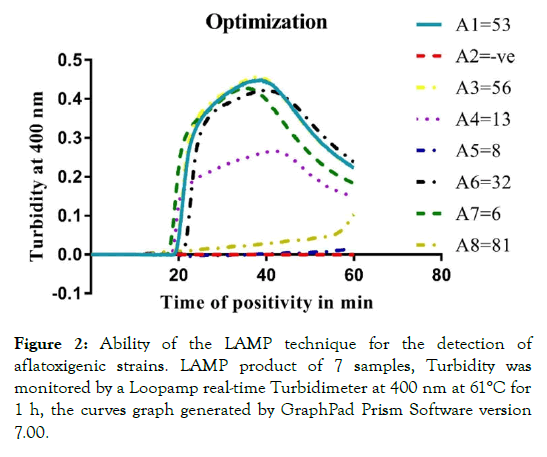
Figure 2: Ability of the LAMP technique for the detection of aflatoxigenic strains. LAMP product of 7 samples, Turbidity was monitored by a Loopamp real-time Turbidimeter at 400 nm at 61°C for 1 h, the curves graph generated by GraphPad Prism Software version 7.00.
To investigate the specificity of the LAMP method, 14 randomly selected samples were tested using the same conditions (Block A and B) and a non-template negative control of reactions was included in each block. The reaction curves showed the positive amplification results in Block A, whereas in block B, 2 amplification results (B4 and B8) produced a negative reaction (Figure 3).
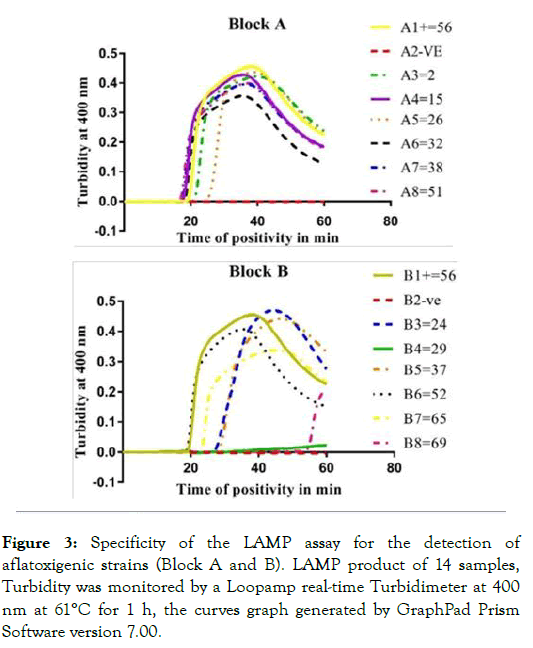
Figure 3: Specificity of the LAMP assay for the detection of aflatoxigenic strains (Block A and B). LAMP product of 14 samples, Turbidity was monitored by a Loopamp real-time Turbidimeter at 400 nm at 61°C for 1 h, the curves graph generated by GraphPad Prism Software version 7.00.
Specificity of the LAMP assay for the detection of aflatoxigenic pathogens was analyzed using genomic DNA isolated from pure cultures of 137 Aspergillus species. Evaluation of the LAMP assay revealed that 98 samples out of 137 genomic DNA were positive of the aflatoxin gene and could be able to produce aflatoxin. To confirm further the detected products, amplicons of the LAMP products were run on agarose gel electrophoresis (Figure 4).
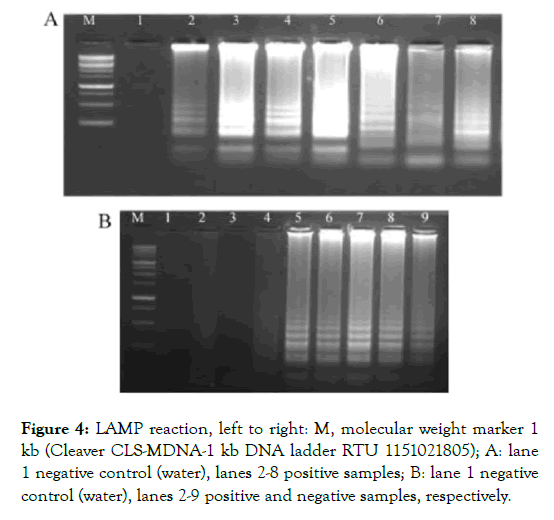
Figure 4: LAMP reaction, left to right: M, molecular weight marker 1 kb (Cleaver CLS-MDNA-1 kb DNA ladder RTU 1151021805); A: lane 1 negative control (water), lanes 2-8 positive samples; B: lane 1 negative control (water), lanes 2-9 positive and negative samples, respectively.
PCR detection
Regarding PCR, 137 samples were screened for the specificity to detect aflatoxigenic species. This method successfully detected 63 out of 137 putative samples aflatoxigenic isolates. This was confirmed by a 250 bp band (Figure 5) of DNA representing nor-1 gene.
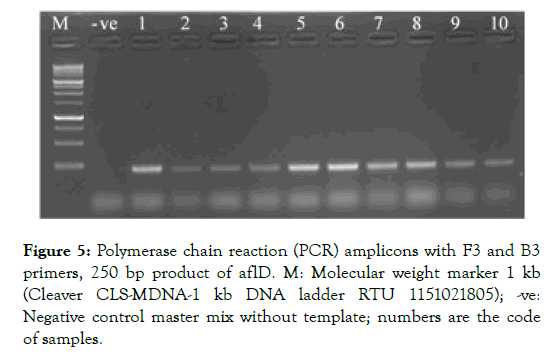
Figure 5: Polymerase chain reaction (PCR) amplicons with F3 and B3 primers, 250 bp product of aflD. M: Molecular weight marker 1 kb (Cleaver CLS-MDNA-1 kb DNA ladder RTU 1151021805); -ve: Negative control master mix without template; numbers are the code of samples.
Application of the LAMP for aflatoxigenic screening demonstrated more amplification efficiency and specificity results than the conventional PCR.
Sensitivity of the LAMP and PCR assays
Sensitivity of the LAMP technique was analyzed using different concentrations of pure genomic DNA of the aflatoxigenic species screened in previous experimentations. After tenth fold serial dilution of DNA (10 μL of pure template into 90 μL of Tris to 10-7 times), positive amplification results were observed with concentrations from 10 to 10-4 μL, whereas examination of amplification curves revealed the limit of the LAMP assay with low concentration (exponential 10-5 to 10-7) under the same conditions (Figure 6A). On the other hand, amplification of the conventional PCR for eight, 10-fold dilution showed no bands of amplified products with concentrations less than 10-2 exponential (Figure 6B). Regarding the findings, these results showed that the LAMP technique could be more efficient and rapid for detecting aflatoxigenic fungal than the PCR-based assays.
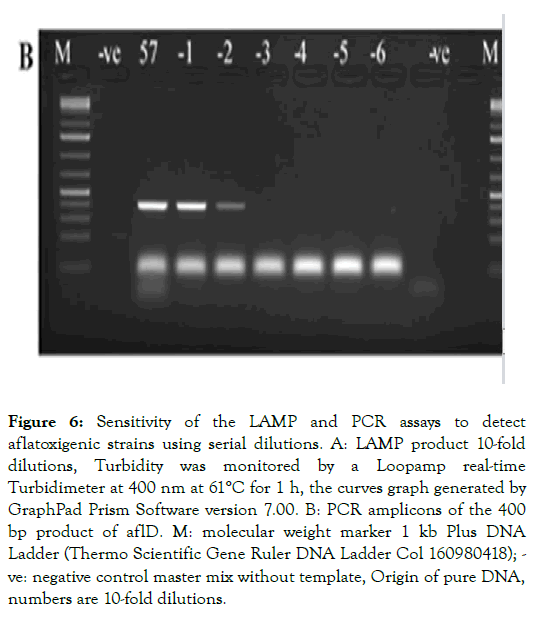
Figure 6: Sensitivity of the LAMP and PCR assays to detect aflatoxigenic strains using serial dilutions. A: LAMP product 10-fold dilutions, Turbidity was monitored by a Loopamp real-time Turbidimeter at 400 nm at 61°C for 1 h, the curves graph generated by GraphPad Prism Software version 7.00. B: PCR amplicons of the 400 bp product of aflD. M: molecular weight marker 1 kb Plus DNA Ladder (Thermo Scientific Gene Ruler DNA Ladder Col 160980418); - ve: negative control master mix without template, Origin of pure DNA, numbers are 10-fold dilutions.
Discussion
The results of isolation showed the appearance of Aspergillus species in all samples incubated, 8 different species being observed with high prevalence. These findings are in accord with several other studies who reported a high prevalence of Aspergillus species contaminants of rice grains from local markets [32-34]. In the current study, the high prevalence of Aspergillus species could be a threat to the consumers towards the contamination of these grains by aflatoxins [35]. Due to the harmful effects of aflatoxins, there is an urgent need to develop a sensitive, rapid, and specific method for the detection of aflatoxigenic pathogens in order to prevent threats by aflatoxins in food.
In our study, the LAMP reaction targeted nor-1 gene DNA for screening aflatoxigenic species from Aspergillus species isolated and compared to the conventional PCR. Two pairs of primers (FIP-BIP and F3-B3) were used for targeting a specific gene interest nor-1 for detecting aflatoxigenic species [24]. In additional, two loop primers (LF and LB) were added to accelerate efficiency of the reaction [36]. We evaluated the practical applicability of the LAMP assay for 137 samples of genomic DNA isolated from Aspergillus species in pure cultures. The positive curve monitored under a Loopamp real-time Turbidimeter could be seen directly through the screen during the LAMP technique [37].
Optimized conditions in this study allowed a complete amplification of targeting gene less than 30 min, amplification curves demonstrated a higher slope (pick) as shown in the Figure 2. Reactions curves of targeting gene started to have positive amplification for aflatoxigenic between 17 to 25 min for the primers and reagents set in this technique. Interestingly, our results demonstrated that the assay can detect aflatoxin producing fungi reliably even under lower concentrations of DNA as shown in serial dilution. This is in line with reports by Cai et al.[38] who demonstrated that even DNA pathogens with low levels can be detected by real-time LAMP. In overall, the assay has shown that the aflatoxigenic species were detected successfully with 71.5% of positive amplification results by the LAMP assay being detected within 30 min of the reaction. These results exhibited that the innovated technique could be very minimally affected by inhibitors during process of the amplification of genomic DNA and reaction can successfully teak place. In contrast, PCR evaluation showed 45.9% of positive amplified products through agarose gel electrophoresis images. This indicates the superiority of the LAMP technique compared to the conventional PCR to decrease during diagnostic procedure due to certain inhibitory substances and the low concentration of the template DNA. These results are in concordance with that recorded by many investigators [32,39-42], which reported that the LAMP assay is specific, sensitive and can detect the pathogens rapidly compared with conventional PCR-based assays. The main advantage of the alternative technique described in the current study is the ability to detect the nor-1 gene fragment of aflatoxigenic strains without pretreatment of genomic DNA, or RNA isolated from Aspergillus. Importantly, positive reactions can be diagnosed within just 30 min and the results can be easily interpreted by Loopamp real-time turbidimeter examination. Moreover, the LAMP method is easier to operate, specific and efficient; it performs under isothermal conditions and can be used by persons with no operating experience.
Conclusions
The current study describes an alternative specific, rapid and reliable method to detect aflatoxigenic species contaminants. We have applied the LAMP technique to detect aflatoxigenic species isolated from rice grains. The study revealed that the LAMP assay succeeded in detecting aflatoxigenic fungal isolated with high specificity (71.5%) and sensitivity (up to eight, 10-fold dilution). Our results demonstrated that the assay is a promising and convenient method, robust and fast for the detection of aflatoxin producers in less time consuming. The LAMP technique excludes the need of sophisticate equipment such as PCR machines, gel electrophoresis or gel image systems for the detection and visualization. Our results will contribute to the body for knowledge on reliable methods that can be used in the detection of aflatoxigenic species of fungi in order to improve strategies, prevent and control fungi pathogens colonization and aflatoxins risks in food and commodities.
Authors Contributions
Youmma Douksouna and Allan Ole Kwallah, conceived this research and designed experiments, Andrew Nyerere, Steven Runo and Zachée Ambang, supervised the research, YD analyzed, interpreted the data and wrote the paper. All authors read and approved the final manuscript.
Funding
This work was funded by the African Union through Pan African University Institute for Basic Sciences, Technology and Innovation (PAUSTI).
Acknowledgments
The authors would like to thank Ronald Tonui, Jemutai Kosgei Lydia who helped and contributed to the research process.
REFERENCES
- Mitchell NJ, Bowers E, Hurburgh C, Wu F. Potential economic losses to the US corn industry from aflatoxin contamination. Food Addit. 2016;33:540-550.
- Lai X, Liu R, Ruan C, Zhang H, Liu C. Occurrence of aflatoxins and ochratoxin A in rice samples from six provinces in China. Food Control. 2015;50:401-404.
- Cotty PJ, Jaime-Garcia R. Influences of climate on aflatoxin producing fungi and aflatoxin contamination. Int J Food Microbiol. 2007;119:109-115.
- Pitt JI. The current role of Aspergillus and Penicillium in human and animal health J Med Vet Mycol. 1994;32:17-32.
- Amaike S, Keller NP. Aspergillus flavus. Annu Rev Phytopathol. 2011;49:107-133.
- Frisvad JC, Hubka V, Ezekiel CN, Hong SB, Nováková A, Chen AJ, et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud Mycol. 2019; 93:1-63.
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447-459.
- Reddy K, Reddy C, Muralidharan K. Detection of Aspergillus spp. and aflatoxin B 1 in rice in India. Food Microbiol. 2006;26:27-31.
- Katsurayama AM, Martins LM, Iamanaka BT, Fungaro MHP, Silva JJ, Frisvad S, et al. Occurrence of Aspergillus section Flavi and aflatoxin in Brasil: From field to market. Int J Food Microbiol. 2018;266:213-221.
- Iqbal SZ, Asi MR, Hanif U, Zuber M, Jinap S. The presence of aflatoxins and ochratoxin a in rice and rice products and evaluation of dietary intake. Food Chem. 2016;210:135-140.
- Commission Regulation (EC) Laying down the Methods of Sampling and Analysis for the Official Control of the Levels of Mycotoxins in Foodstuffs; Official Journal of the European Union. Brussels, Belgium. 2005.
- Reports IWG Mycotoxin Control in Low and Middle-Income Countries; International Agency for Research on Cancer. Lyon, France. 2015.
- Gemeda N, Woldeamanuel Y, Asrat D, Debela A, Lemma H. Assessment of Aflatoxigenic Aspergillus Species in Food Commodities from Local Market of Addis Ababa. Research. 2014.
- Lee J, Her JY, Lee KG. Reduction of aflatoxins (B1, B2, G1, and G2) in soybean-based model systems. Food Chem. 2015;189:45-51.
- Ruadrew S, Craft J, Aidoo K. Occurrence of toxigenic Aspergillus spp. and aflatoxins in selected food commodities of Asian origin sourced in the West of Scotland. Food Chem Toxicol. 2013;55:653-658.
- Naseer R, Sultana B, Khan M, Naseer D, Nigam P. Utilization of waste fruit-peels to inhibit aflatoxins synthesis by Aspergillus flavus: A biotreatment of rice for safer storage. Bioresour Technol. 2014;172:423-428.
- Tola M, Kebede B. Occurrence, importance and control of mycotoxins Bioscience Environment and Agriculture Environmental Studies & Management Food Science and Technology. Cogent Food Agric. 2016;2:1191103.
- Suanthie Y, Cousin MA, Woloshuk CP. Multiplex real-time PCR for detection and quantification of mycotoxigenic Aspergillus, Penicillium and Fusarium. J Stored Prod Res. 2009;45:139-145.
- Rodríguez A, Rodríguez M, Luque MI, Martín A, Córdoba JJ. Real-time PCR assays for detection and quantification of aflatoxin-producing molds in foods. Food Microbiol. 2012;31:89-99.
- Sardiñas N, Vázquez C, Gil-Serna J, González-Jaén MT, Patiño B. Specific detection and quantification of Aspergillus flavus and Aspergillus parasiticus in wheat flour by SYBR® Green quantitative PCR. Int J Food Microbiol. 2011;145:121-125.
- Mangal M, Khan F, Bansal S, Oberoi H. Validation of PCR based detection system for aflatoxin producing molds. Indian J Exp Biol. 2016;54: 472-476.
- Tombelli S, Mascini M, Scherm B, Battacone G, Migheli Q. DNA biosensors for the detection of aflatoxin producing Aspergillus flavus and A. parasiticus. Mon Chem Mon. 2009;140:901-907.
- Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors-occurrence, properties and removal. J Appl Microbiol. 2012;113:1014-1026.
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28-63.
- Ludwig N, Julia B, Sihem F, Marta HT, Rudi FV. LAMP-based group specific detection of aflatoxin producers within Aspergillus section Flavi in food raw materials, spices, and dried fruit using neutral red for visible-light signal detection. Int J of Food Microbiology. 2018;266:241-250.
- Jie L Rudi FV, Ludwig N. Rapid detection of aflatoxin producing fungi in food by real-time quantitative loop-mediated isothermal amplification. Food Microbiology. 2014;44:42-148
- Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223-229.
- Pitt JI, Hocking AD. Fungi and food spoilage. Blackie Academic and Professional, Cambridge, UK. 1997.
- Atlas RM. Handbook of Microbiological Media; CRC Press: Boca Raton, FL, USA. 2010.
- Pitt JI, Hocking AD. Fungi and Food Spoilage, 3rd ed, Springer: New York, NY, USA.20009.
- Varga J, Frisvad JC, Samson RA. Two new aflatoxin producing species and an overview of Aspergillus section. Flavi Stud Mycol. 2011;69:57-80.
- Shanakht H, Shahid AA, Ali SW. Characterization of fungal microbiota on rice grains from local markets of Lahore. Journal of Hygienic Engineering and Design. 2014;37:35-40.
- Renu K, Agarwal MK, Bhagayavant SS, Verma P, Nagar DP. Detection of Aspergillus flavus using PCR method from fungus infested food grains collected from local market. Ann Plant Sci. 2018;7:2073-2077.
- Ibrahim F, Jalal H, Khan AB, Asghar MA, Iqbal J, Aftab A, et al. Prevalence of Aflatoxigenic Aspergillus in food and feed samples from Karachi. J Infect Mol Biol Preval. 2016;4:1-8.
- Aydin A, Aksu H, Gunsen U. Mycotoxin levels and incidence of mould in Turkish rice. Environ Monit Assess. 2010;10:1661-1688.
- Monica NI, Rathinasabapathi P, Ramya M. Development of real-time loop-mediated isothermal amplification (Real Amp) method for sensitive and rapid detection of pathogenic and nonpathogenic Leptospira. Letters in Applied Microbiology. 2019;68:196-203.
- Karthikeyan K, Sharma A, Mekata T, Itami T, Sudhakaran R. Rapid and sensitive real-time loop medicated isothermal amplification for the detection of Enterocytozoon hepatopenaei of shrimp. Aquaculture. 2017;481:119-123.
- Cai SX, Kong FD, Xu SF, Yao CL. Real-time loop-mediated isothermal amplification for rapid detection of Enterocytozoon hepatopenaei. Peer J. 2018;6:e5993.
- Xue Y, Mohamed NA, Yong Z, Ai-Fang Z, Hao-Yu Z, Gu CY, et al. Rapid Detection of Ustilaginoidea virens from Rice using Loop-Mediated Isothermal Amplification Assay. Changsha 410004, China, Plant Disease. 2018;1741-1747.
- Aongart M, Nipa T, Amonrattana R, Ruenruetai U, Supaluk P, Sukhontha S, et al. Development of a loop-mediated isothermal amplification technique and comparison with quantitative real-time PCR for the rapid visual detection of canine neosporosis. Parasites & Vectors. 2017;10:394.
- Al-Sheikh HM. LAMP-PCR detection of ochratoxigenic Aspergillus species collected from peanut kernel. Genetics and Molecular Research. 2014;14:634-644.
- Mohammad AA, Seyed MHD, Aboubakr M, Zahra E, Mehdi AO. Development and Application of Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Fusarium Oxysporum f. Sp. Lycopersici. J Plant Pathol Microb. 2013;4:5.
Citation: Douksouna Y, Kwallah AO, Nyerere A, Runo S, Ambang Z (2020) Application and Evaluation of the Loop-Mediated Isothermal Amplification assay for the Detection of Aflatoxigenic Fungi Contaminants of Rice Grains in Kenya. Plant Pathol Microbiol. 11:491. doi: 10.35248/2157-7471.20.11.491.
Copyright: © 2020 Douksouna Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.

