Indexed In
- Open J Gate
- Genamics JournalSeek
- China National Knowledge Infrastructure (CNKI)
- Ulrich's Periodicals Directory
- RefSeek
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Publons
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research - (2024) Volume 14, Issue 2
Antibacterial Susceptibility of Bacteria Isolated from Wound Swab of Patient Receiving Medication in University Teaching Hospital, Gwagwalada, Abuja, Nigeria
Jimba Rai Amos*, Egbenoma Aigboeghian, Aigboeghian Preciousgift and OO OmolehinReceived: 20-Mar-2024, Manuscript No. CPECR-24-25184; Editor assigned: 25-Mar-2024, Pre QC No. CPECR-24-25184 (PQ); Reviewed: 08-Apr-2024, QC No. CPECR-24-25184; Revised: 15-Apr-2024, Manuscript No. CPECR-24-25184 (R); Published: 22-Apr-2024, DOI: 10.35248/2161-1459.24.14.419
Abstract
This research examined the antibacterial sensitivity of bacteria isolated from wound infection. Ten samples were collected using swab stick. From the isolates of this research identified both Gram positive and Gram negative bacteria; these include Staphylococcus aureus, Staphylococcus epidermidis, Enterococci, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Enterobacter species. These organisms are of public health importance. Susceptibility outcome revealed that Ofloxacillin and ciprofloxacin were effective against Gram positive bacteria. Amikacin, Ciprofloxacin, Ceftriaxone and Norflaxacin were effective against Gram negative bacteria hence, the study indicate that Gram negative bacteria are more susceptible to antibiotic than Gram positive bacteria. Antimicrobial Multi-Resistant (AMR) was higher among Gram positive bacteria compared to Gram negative bacteria. Treatment of wound infection remains a significant concern for surgeons and physicians in a health care facility. The problem has been magnified due to the unrestrained and rapidly spreading resistance to the available array of antimicrobial agents. The study therefore, recommends multi-drug administration to patient suffering from wound infection, either caused by burn, accident or of any kind.
Keywords
Antimicrobial; Sensitivity; Microorganisms; Wound; Isolates; Wound infection
Introduction
Over years, wound infections are one of the most common hospital acquired infections and are an important cause of morbidity and account for 70%-80% mortality, which can be caused by different groups of microorganisms like bacteria, fungi and protozoa. However, different microorganisms can exist in polymicrobial communities especially in the margins of wounds and in chronic wounds. The infecting microorganism may belong to aerobic as well as anaerobic group. Most commonly isolated aerobic microorganism include Staphylococcus aureus, Coagulase-negative staphylococci, Enterococci, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Enterobacter species, Proteus mirabilis, Candida albicans and Acinetobacter.
Wound infections have been a problem and are the field of medicine for a long time. The presence of foreign materials increases the risk of serious infection even with relatively small bacterial inoculums. Advances in control of infections have not completely eradicated this problem because of development of drug resistance. The widespread uses of antibiotics, together with the length of time over which they have been available have led to major problems of resistant organisms contributing to morbidity and mortality. Hence, antimicrobial resistance can increase complications and costs associated with procedures and treatment.
Wound infections are one of the most important and potentially serious complications that occur in the acute period following injury which are subsequently colonized with microorganisms, including gram-positive bacteria, gram-negative bacteria and yeasts, which derived from the host’s normal flora (gastrointestinal flora, upper respiratory flora) and from the hospital environment. Microorganisms may also be transferred to a patient’s skin surface via contact with contaminated external environmental surfaces, water, fomites, air, hydrotherapy treatment, and the soiled hands of health care workers. The risk of invasive wound infection is influenced by the extent and depth of the burn injury, various host factors, and the quantity and virulence of the microbial flora colonizing the wound.
The common burn wound pathogens are Pseudomonas aeruginosa, Klebsiella spp. and Staphylococcus aureus, which produce a number of virulence factors that are important in the pathogenesis of invasive infection. Wound especial those cause by burns are the most devastating of injuries and burn patients may suffer from their complications for the rest of their lives. In spite of the recent advances in burn care, still high mortality and significant morbidity is noted in terms of complications associated with burn wounds. In developing countries, more than 90% of fatal fire-related burns occur, over half of which alone occur in South East Asia. Data collected from different areas of the world shows that 75% of deaths in burn patients are due to infections of burn wounds. More than 10,000 African die every year from infections associated with burns. This implied that wound infections caused by burn are serious case in life threatened patient which this study tends to look for sensitive antibiotic for. Hence, the skin barrier is the natural guard to prevent the entry of pathogenic organisms inside the body. Destruction of skin barrier provides favourable entry site for the bacteria to invade and grow. Wound sepsis remains the most dangerous out-come in patients who have suffered major burn injuries and leads to overwhelming mortality among patients with extensive burn wounds.
According to research conducted by Bayeh et al. [1], stated that burn wounds will almost inevitably be colonized by microorganisms within 24 h to 48 h and this may remain as localized infection. In addition, there may bacteremia or septicemia and metastatic infections may develop at other body sites. This means that Bacteremia is a common cause of fatality in severe wound especially those cause as a result of burns patients and may occur any time from the first day until the point when all the wounds have entirely healed. Other major factors responsible for mortality in burn victims are fluid and protein loss, pulmonary edema and pneumonia.
The presence of large areas of devitalized, necrotic tissue, coupled with the profound immune-suppression that usually follows major burn injuries, sets the stage for rapid microbial proliferation in the wounds; when microbes invade adjacent, previously viable tissues, invasive burn wound sepsis is developed. Topical antimicrobial drugs probably have only a limited role in preventing wound sepsis, and organisms now frequently emerge that are resistant to commonly used topical agents. Wound caused by injuries provide favourable sites for colonization and growth of microorganisms acquired from body’s own indigenous flora, flora of the hospital staff or from the environment surrounding these patients [2]. Immediately after injury, burned tissues contain no microorganisms. Within 24 h microbial colonization occurs. Gram Positive organisms grow first, followed by colonization of Gram negative species.
The predicted problem of mortality rises by 50% when Gram-negative organisms are associated with bacteremia in burn patients. Cost associated with the management of burn patients is also too high; which could also contribute to incomplete and hence ineffective management of burn patients, contributing to emergence of resistance in pathogens [3].
Increasing antibiotic use and misuse in humans, animals, agriculture, along with poor infection control strategies and some other factors have been reported for increasing resistance to commonly used antimicrobial agents [3]. The current study will be conducted to determine the antimicrobial susceptibility of some common bacterial isolates from wounds swap in University of Abuja Teaching Hospital to help policy makers in formulation of strategies for rational and effective use of antimicrobial agents. This might help in the control of spread of antibiotic resistance genes in the community and in the reduction of morbidity and mortality associated with better management of wound patients.
Antibiotic prophylaxis is indicated in situations or wounds at high risk to become infected such as: contaminated wounds, penetrating wounds, abdominal trauma, compound fractures, lacerations greater than 5 cm, wounds with devitalized tissue, and high risk anatomical sites such as hand or foot, etc. According to Khameneh et al. [4], Andhoga et al. [5], Anguzu et al. [6], recommended that prophylaxis consists of penicillin G and metronidazole given once. These indications apply for injuries which may or may not require surgical intervention. For injuries requiring surgical intervention, antibiotic prophylaxis is also indicated and should be administered prior to surgery, within the 2 hour period before the skin is cut.
However, antibiotics play an important role in the treatment of bacterial infections. Several reports indicate an increasing rate of bacterial resistance. However, this present study shall be significance to the following people: Clinicians; the ministry of Health; the government worldwide and the patient. The aim of the study was to determine susceptibility patterns of microorganisms isolated from wound swab of patient in University of Abuja Teaching Hospital, Gwagwalada Abuja.
Materials and Methods
Collection of samples and identification
Samples were collected from the ten (10) patients with complaints of delayed and non-healing wound infection. The wound samples will be collected by using a sterile cotton swab or swab stick, the inner surface of the infected area was swabbed gently and then the swabs was transported to the microbiology laboratory for examination and sensitivity screening.
Preparation and characterization of media
About 19 g of MacCkey agar was dissolved in 38 ml distilled water and then sterilized in an autoclave at 121°C for 15 minutes. After autoclaving, the agar medium was allowed to cool to 37°C before pouring into the sterile Petri dishes to avoid condensation that may course contamination and necessary precaution was taken to avoid other contamination.
Preparation of the inoculums
A loopful of each test organism was prepared using striking method from their respective agar and subculture in the sterile Petri dishes and incubated at 37°C for 24 hours. Pure cultures of the test organisms were used for sensitivity screening.
Bacteriology
In the laboratory, each sample was inoculated on MacConkey agar, Nutrient agar and Blood agar. The inoculums on the plate were streaked out for discrete colonies with a sterile wire loop. The culture plates was incubated at 37°C for 24 hours and observed for growth through the formation of colonies. A the bacteria will be isolated and identified using morphological, microscopy and biochemical tests following standard procedures described by Sharma et al. [7], shall be employed.
Antibiotic susceptibility testing
Antimicrobial susceptibility test were carried out on isolated and identified colonies of Gram-negative bacteria using commercially prepared antibiotic disk (Span diagnostics) on Nutrient agar plates by the disk diffusion method, according to the Central Laboratory Standards Institute (CLSI) guidelines. Antibiotics used in our study were Ticarcillin/Clavulanic acid, Meropenem, Levofloxacin, Moxifloxacin, Cefprozil, Cefirome, Ceftizoxime, Cefpodoxime, Cefoperazone or Sulbactam, Sparfloxacin, Pipercillinor Tazobactum, Gatifloxacin, Imipenem or Cilastatin, amikacin (10 μg), gentamicin (10 μg) and Tobramycin, Amoxicillin or clavulanic acid (30 μg). While for Gram-positive organism additional antibiotics used different from the ones mentioned above are clindamycin (5 μg), oxacillin (1 μg) and erythromycin (10 μg).
Data analysis
The statistic method that was used for analysing the result of this study was frequency, mean and Chi-square test.
Biochemical tests for the isolates
Gram stain: A pure colony was spread and fixed on the slide by drying using a Bunsen burner flame. The slide was allowed to cool, and then flooded with crystal violet solution for 30 sec, followed with Grams iodine solution for 1 min, followed by draining excess iodine by decolorizing using acetone for at least 10 sec and then washed with water. Counter staining was done using Basic fuchsine and allowed to stand for 30 seconds. This was followed by washing the slide and dried in the air. The slide was observed under light microscopy at 40X. Short rods that stained red pink were considered gram negative.
Catalase test: Catalase test was used to determine whether or not a microorganism produces catalase. A loopful of the culture suspension or tiny inoculums on the clean-grease-free slide. Emulsify the culture with a loopful of freshly prepared hydrogen-peroxide (H2O2). Effervescence, indicating free oxygen liberation bubbles signify the presence of catalase in the Hydrogen-peroxide, while the absence of bubbles is an indication of catalase negative.
Indole production: Two to five pure colonies were inoculated using a sterile wire loop in 2 ml of peptone water in bijous bottles and incubated overnight at 35°C. 0.5 ml of Kovac’s reagent was added and examined after 1 minute. Presence of rose red colour on upper layer was considered positive (+), while absence of rose red or pale colour was considered negative (-).
Coagulase test: In each bijous bottle, 2.5 ml of Methyl red-Voges Proskauer broth was added and inoculated with pure colonies of test organisms. The bijous bottles were then incubated at 35°C for 48 h, followed by addition of 0.6 ml or 6 drops of VP reagent A (α-naphthanol 34 solution), then 0.2 ml (2 drops) of VP reagent B (40% KOH). The bijous bottles were shaken and allowed to stand for 15 minutes. Pink red colour (reddish pink) of the broth culture in the bijous bottles was considered positive (+), while colourless (pale) were considered negative (-).
Methyl red test: Five millilitres of Methyl red-Voges Proskauer broth was distributed in bijous bottles and inoculated with pure colonies of test organisms. The bijous bottles were incubated at 35°C for 48 h, followed by addition of 0.5 ml or 5 drops of methyl red and observed for colour change. The bijous bottles with red colour were considered positive (+), while those which developed yellow colour were considered negative (-).
Simmons citrate: Simmons Citrate agar slants in bijous bottles were stabbed using a sterile wire loop and incubated for 48 h at 35°C. Positive (+) growth for example citrate utilization produce an alkaline reaction and the medium change colour from green to blue, while no colour change (no citrate utilization) was considered negative.
Urease test: Two millilitres of Urea broth in bijous bottles were inoculated with single colonies of organism and incubated for 5-6 h at 37°C in a water bath. Two controls were used, a negative control containing Urea broth base only and positive control containing Proteus aureus standard organism. All bijous bottles in which colour changed to pink were considered positive (+), while those that had no colour change were considered negative.
Results
The results of this research work (Table 1), reveals the bacteria involved in the wound infection and their resistance to the selected antibiotics. From the four sampling sites collected, about three genus of Gram negative bacteria were isolated; these include Staphylococcus aureus, Klebsiella pneumonia, Pseudomonas aeruginosa, Staphylococcus epidermidis, Acinetobacter species and Proteus vulgaris. These organisms are of public health importance. They all showed multiple resistances to above mentioned antimicrobial agents. The results for biochemical, sensitivity testing was shown in the Table 1.
| Isolated code | Cat | Co | Ind | Ox | Ci | Met | Man |
|---|---|---|---|---|---|---|---|
| Staphylococcus aureus | _ | + | + | S | + | + | + |
| Pseudomonas aeruginosa | + | _ | _ | S | _ | + | _ |
| Klebsiella pneumonia | + | _ | _ | R | + | _ | + |
| Staphylococcus epidermidis | + | _ | _ | NT | + | + | _ |
Note: (+): Positive reaction; (-): Negative reaction; (+SI): Slow positive reaction; V: Variable; S: Sensitive; Cat: Catalase; Co: Coagulase; Met: Methyl red; Glu: Glucose; Ind: Indole test; Ci: Citrate; Manni: Mannitol; NT: Not Tested; Ox: Oxidase test.
Table 1: The biochemical characteristics of the isolated bacteria from wound swabs.
Table 2, revealed the number of colonies in each plate of culture; Staphylococcus aureus 23 (33%), Pseudomonas aeruginosa 20 (29%), Klebsiella pneumonia 9 (13%), Escherichia coli 10 (14%) and Staphylococcus epidermidis 8 (11%). Hence, it can be deduce that Staphylococcus aureus had the highest colonies while the least was Staphylococcus epidermidis.
| Bacterial | Number of isolate | Percentage |
|---|---|---|
| Staphylococcus aureus | 23 | 33% |
| Pseudomonas aeruginosa | 20 | 29% |
| Klebsiella pneumoniae | 9 | 13% |
| Escherichia coli | 10 | 14% |
| Staphylococcus epidermidis | 8 | 11% |
| Total | 70 | 100 |
Table 2: The prevalence of bacterial isolates from wound infection.
Figure 1, using duplicated plates (two culture plate) on the measurement of zone of Inhibition of Gram positive organisms, mental rule was used for the measurement of zone of inhibition. Amikacin, Ofloxacillin and Vancomycin were resistant to Staphylococcus aureus. Gentamicin, Cotrimoxazole and Piperacillin were intermediate on Staphylococcus aureus while Ciprofloxacin was susceptible to Staphylococcus aureus. Hence, it can be deduced that antibiotic has less reactivity on Staphylococcus aureus compare to Staphylococcus epidermidis.
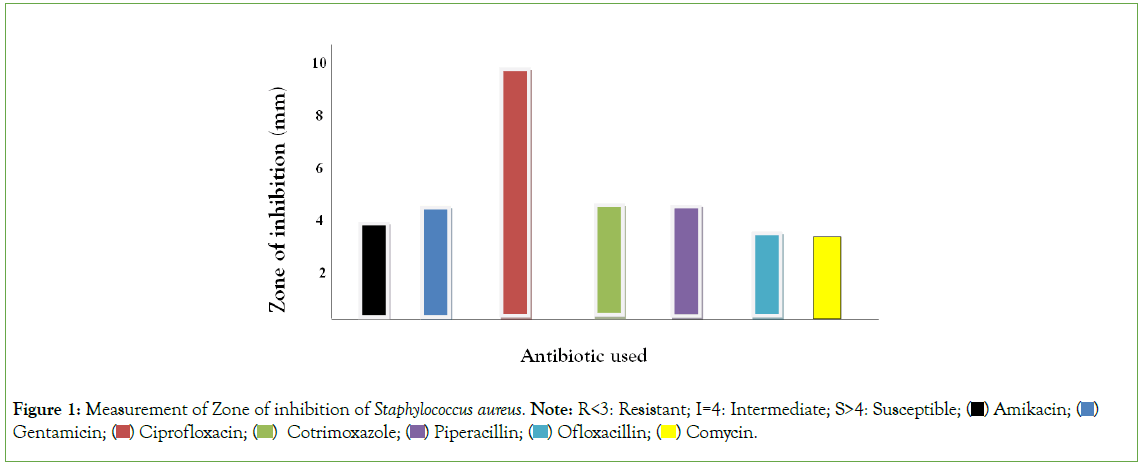
Figure 1: Measurement of Zone of inhibition of Staphylococcus aureus. Note: R<3: Resistant; I=4: Intermediate; S>4: Susceptible;  Gentamicin;
Gentamicin;  Comycin.
Comycin.
Figure 2, using duplicated plates (two culture plate) on the measurement of zone of Inhibition of Gram positive organisms, mental rule was used for the measurement of zone of inhibition. Gentamicin, Cotrimoxazole, Piperacillin and Vancomycin was resistant to Staphylococcus epidermidis. Amikacin was intermediate on Staphylococcus epidermidis while Ofloxacin, and Ciprofloxacin were susceptible to Staphylococcus epidermidis. Hence, it can be deduced that antibiotic has moderate reactivity on Staphylococcus epidermidis compare to Staphylococcus aureus.

Figure 2: Measurement of Zone of inhibition of Staphylococcus epidermidis. Note: R<3: Resistant; I=4: Intermediate; S>4: Susceptible. Note:  mikacin;
mikacin;  Vancomycin.
Vancomycin.
Figure 3, using duplicated plates (two culture plate) on the measurement of zone of Inhibition of Gram negative organisms, mental rule was used for the measurement of zone of inhibition. Gentamicin, Ciprofloxacin and Cephalexin Amoxicillin, was resistant to Klebsiella pneumoniae while Norfloxacin, Ofloxacin, Ceftriaxone, Ciprofloxacin and Amikacin were susceptible to Klebsiella pneumonia but of the antibiotic has intermediate reaction on Klebsiella pneumoniae. Hence, it can be deduced that antibiotic is highly susceptible to Klebsiella pneumoniae.
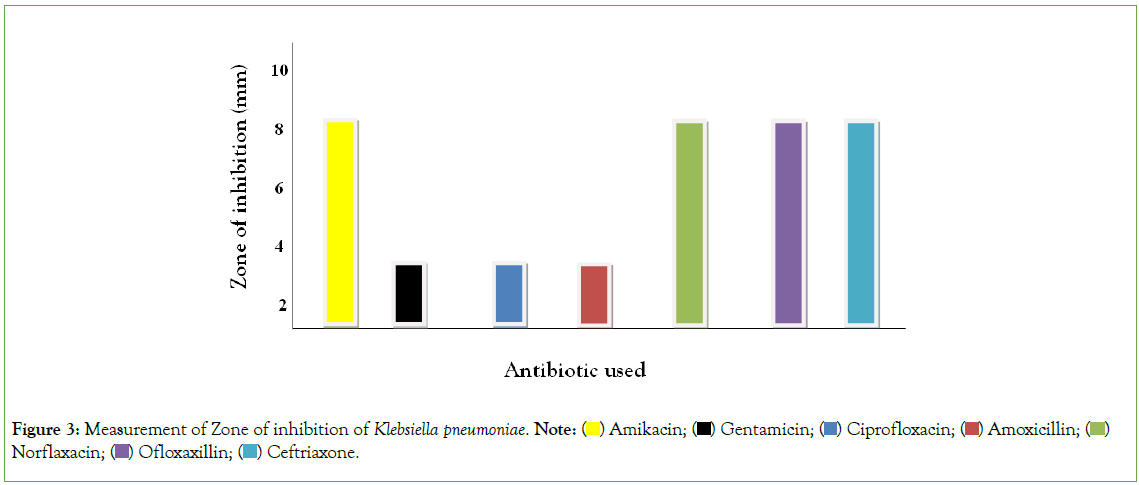
Figure 3: Measurement of Zone of inhibition of Klebsiella pneumoniae. Note:  Norflaxacin;
Norflaxacin;  Ceftriaxone.
Ceftriaxone.
Figure 4, using duplicated plates (two culture plate) on the measurement of zone of Inhibition of Gram negative organisms, mental rule was used for the measurement of zone of inhibition. Gentamicin, Norfloxacin, and Cephalexin Amoxicillin, was resistant to Escherichia coli Ofloxacin was intermediate on Escherichia coli while Ceftriaxone, Ciprofloxacin and Amikacin was susceptible to Escherichia coli. Hence, it can be deduced antibiotic has moderate reaction or susceptible to Escherichia coli.

Figure 4: Measurement of Zone of inhibition of Escherichia coli. Note:  Norflaxacin;
Norflaxacin;  Ceftriaxone.
Ceftriaxone.
Figure 5, using duplicated plates (two culture plate) on the measurement of zone of Inhibition of Gram negative organisms, mental rule was used for the measurement of zone of inhibition. Gentamicin, Ofloxacin, Ceftriaxone and Cephalexin, was resistant to Citrobacter spp. Amikacin and Ciprofloxacin was intermediate on Citrobacter spp while Amoxicillin and Norfloxacin was susceptible to Citrobacter spp. Hence, it can be deduced antibiotic is less susceptible to Citrobacter spp.
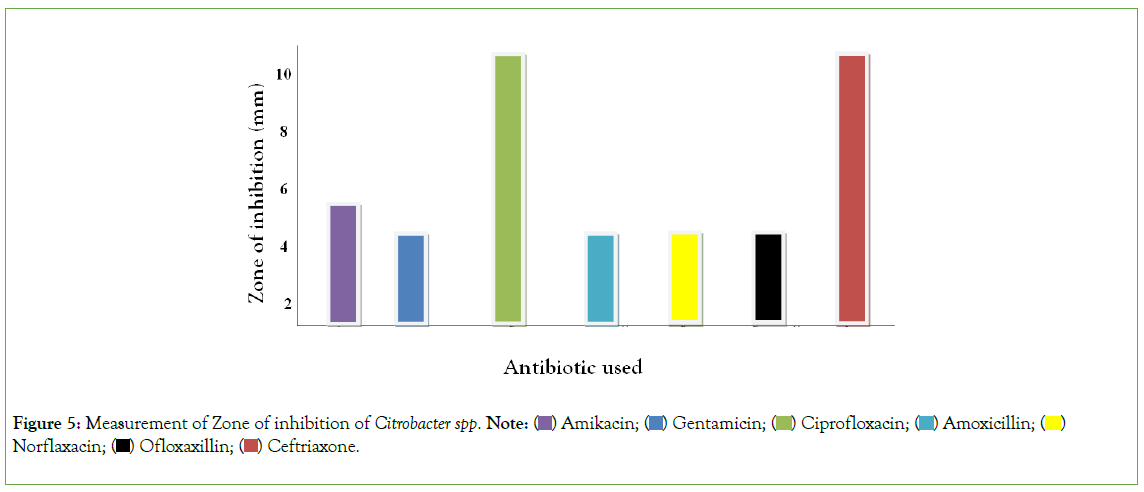
Figure 5: Measurement of Zone of inhibition of Citrobacter spp. Note:  Norflaxacin;
Norflaxacin;  Ceftriaxone.
Ceftriaxone.
Figure 6, using duplicated plates (two culture plate) on the measurement of zone of Inhibition of Gram negative organisms, mental rule was used for the measurement of zone of inhibition. Amikacin, Cephalexin, Ciprofloxacin and Amoxicillin was resistant to Pseudomonas aeruginosa. Norfloxacin and Ofloxacin was intermediate on Pseudomonas aeruginosa while Gentamicin and Ceftriaxone was susceptible to Pseudomonas aeruginosa. Hence, it can be deduced antibiotic is less susceptible to Pseudomonas aeruginosa.
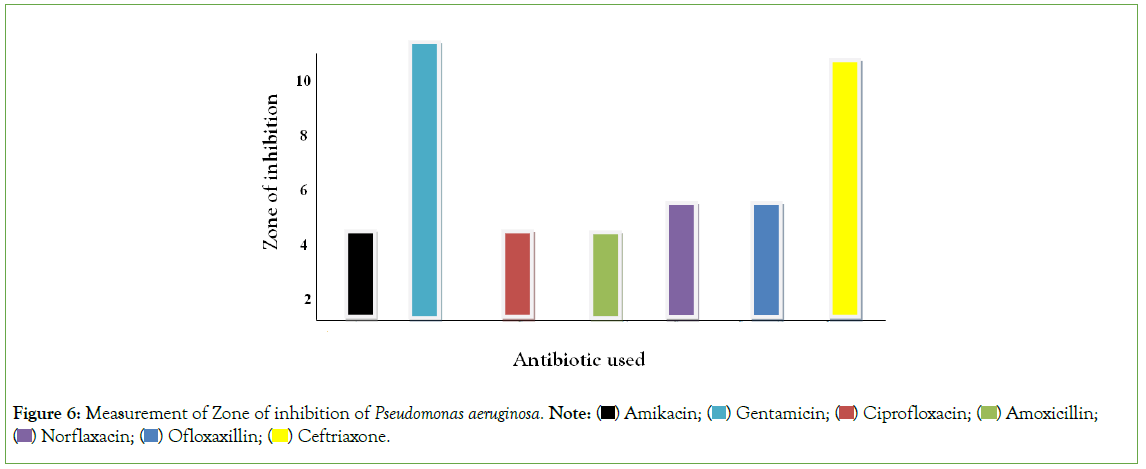
Figure 6: Measurement of Zone of inhibition of Pseudomonas aeruginosa. Note:  Amoxicillin;
Amoxicillin;  Ceftriaxone.
Ceftriaxone.
Table 3, showed the o-value for both treatment and zones of inhibition of both Gram positive and Gram negative isolates which from the calculated value had greater value than 0.05 at 5%. Hence, we accept the null hypothesis that says Gram negative bacteria are more susceptible to antibiotic than Gram positive bacteria.
| Source | Type II sum of squares | Df | Mean square | F | p-value |
|---|---|---|---|---|---|
| Corrected model | 75.600 | 3 | 25.200 | 0.472 | 0.704 |
| Intercept | 1153.200 | 1 | 1153.200 | 21.614 | 0.000 |
| Both org. | 58.800 | 1 | 58.800 | 1.102 | 0.303 |
| Colonies | 16.800 | 2 | 8.400 | 0.157 | 0.855 |
| Error | 1387.200 | 26 | 53.354 | ||
| Total | 2616.000 | 30 | |||
| Corrected total | 1462.800 | 29 |
Note: Chi-Squared=0.052 (Adjusted R Squared=-0.058).
Table 3: Test for hypothesis.
Discussion
Management and treatment of wound infections remain a significant concern for surgeons and physicians in a health care facility. The problem has been magnified due to the unrestrained and rapidly spreading resistance to the available array of antimicrobial agents, the only choice with us to treat the infections. In-patients face additional exposure to hospital acquired infections due to longer stays. Data on the bacterial populations are very limited especially involving wound infections. The selection of patients was restricted to those admitted in the post-operative surgical wards after undergoing various surgeries as the infection rates are highest in the surgical wards among the clinical departments. 56 (58.33%) were Gram negative isolates involved in causing post-operative wound infections. Similar observations have been reported from Nigeria.
This could be attributed to be acquired from patient’s normal endogenous microbial fecal flora. The presence of enteric organisms probably resulted in subsequent sepsis. E. coli 24 (42.9%) was the commonest gram negative bacteria isolated. E. coli invasion of the wound is a clear case of poor hospital hygiene, just like other implicated organisms which are frequent agents of nosocomial infections. S. aureus 36 (37.5%) was the single predominant gram positive bacterial isolate obtained. Several reports by CDCP [8], stated that S. aureus as the predominant isolate involved in causing resistance on wound infection.
Susceptibility outcome revealed that ciprofloxacin and Ofloxacillin was the most effective antibiotic against the gram positive bacteria. Amikacin, Ciprofloxacin, Ceftriaxone and Norflaxacin were effective against Gram negative bacteria hence, the study indicate that Gram negative bacteria are more susceptible to antibiotic than Gram positive bacteria.
This suggests a very high resistance gene pool due perhaps to gross misuse, overuse and inappropriate use of the antibacterial agents. The pattern is best understood in terms of selective pressure exerted on the organisms based on the current antibiotics use. Fluoro-quinolones and aminoglycosides are being more frequently prescribed in our settings. Hospitals provide an environment conducive to the spread of resistant organisms among population. Additionally, higher multidrug resistance frequencies in a hospitalized population with intense exposure to antibiotics had been reported. Limitations of the study being that anaerobic bacteria profile and fungal cultures were not done on wound swabs obtained from post-operative wound infection. A continuous monitoring and update studies on the local microbial isolates are an essential and mandatory requirement for a better management and treatment of post-operative wound infections. This would be supplemented with proper infection prevention and control measures and a sound antibiotic policy. This would result in better patient care, safety and health care outcomes.
Recommendations
• The study recommended multi-drug administration to patient suffering from wound infection.
• Similarly, any patient suffering from unhealing/long healing wound should be isolated or quarantine in order to reduce the risk of nosocomial infection because most of the isolated bacterial were nosocomial bacteria.
Conclusion
From the research of the study, both gram positive and gram negative organisms were found dominant in the aetiology of sepsis. Continuous clinical and microbiological surveillance leading to quick detection of aetiological agent(s), appropriate antimicrobial therapy, care for nutrition and early wound cover all factors that can collectively help to reduce mortality and may result in a better outcome. Establishment of burn units in other hospitals situated in other parts can reduce the burden/overcrowding in this unit and will help in the reduction of nosocomial infections.
References
- Bayeh Abera BA, Mulugeta Kibret MK. Bacteriology and antimicrobial susceptibility of otitis media at Dessie Regional Health Research Laboratory, Ethiopia. Ethiop J Health Dev. 2011;25(2):161-167.
- Applegreen BA. The occurrence of illnesses in the workplace and their probable influence on productivity. Doctoral dissertation, North-West University (South Africa). 2002.
- Tayfour MA, Al-Ghamdi SM, Al-Ghamdi AS. Surgical wound infections in King Fahad Hospital at Al-Baha. Saudi Med J. 2005;26(8):1305.
[Google Scholar] [PubMed]
- Khameneh ZR, Afshar AT. Antimicrobial susceptibility pattern of urinary tract pathogens. Saudi J Kidney Dis Transpl. 2009;20(2):251-253.
[Google Scholar] [PubMed]
- Andhoga J, Macharia AG, Maikuma IR, Awnyonyi ZS, Ayumba BR, Kakai R. Aerobic pathogenic bacteria in post-operative wounds at Moi Teaching and Referral Hospital. East Afr Med J. 2002;79(12):640-644.
[Crossref] [Google Scholar] [PubMed]
- Anguzu JR, Olila D. Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda. Afr Health Sci. 2007;7(3).
- Sharma M, Schmid M, Rothballer M, Hause G, Zuccaro A, Imani J, et al. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell Microbiol. 2008;10(11):2235-2246.
[Crossref] [Google Scholar] [PubMed]
- Centers for Disease Control and Prevention. Staphylococcus aureus resistant to vancomycin--United States, 2002. MMWR Morb Mortal Wkly Rep. 2002;51(26):565-567.
[Google Scholar] [PubMed]
Citation: Amos JR, Aigboeghian E, Preciousgift A, Omolehin OO (2024) Antibacterial Susceptibility of Bacteria Isolated from Wound Swab of Patient Receiving Medication in University Teaching Hospital, Gwagwalada Abuja-Nigeria. J Clin Exp Pharmacol. 14:419.
Copyright: © 2024 Amos JR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.