Indexed In
- Open J Gate
- Genamics JournalSeek
- Academic Keys
- ResearchBible
- Cosmos IF
- Access to Global Online Research in Agriculture (AGORA)
- Electronic Journals Library
- RefSeek
- Directory of Research Journal Indexing (DRJI)
- Hamdard University
- EBSCO A-Z
- OCLC- WorldCat
- Scholarsteer
- SWB online catalog
- Virtual Library of Biology (vifabio)
- Publons
- Geneva Foundation for Medical Education and Research
- Euro Pub
- Google Scholar
Useful Links
Share This Page
Journal Flyer

Open Access Journals
- Agri and Aquaculture
- Biochemistry
- Bioinformatics & Systems Biology
- Business & Management
- Chemistry
- Clinical Sciences
- Engineering
- Food & Nutrition
- General Science
- Genetics & Molecular Biology
- Immunology & Microbiology
- Medical Sciences
- Neuroscience & Psychology
- Nursing & Health Care
- Pharmaceutical Sciences
Research Article - (2024) Volume 13, Issue 1
An Optimized dsRNA Production Protocol from Bacterial Cultures: A Case for Fusarium graminearum Genes
Binod Gyawali1, Houshang Alizadeh2 and Mohsen Mohammadi1*2Department of Agronomy and Plant Breeding, University of Tehran, Tehran, Iran
Received: 07-Mar-2024, Manuscript No. AGT-24-25095; Editor assigned: 11-Apr-2024, Pre QC No. AGT-24-25095 (PQ); Reviewed: 25-Mar-2024, QC No. AGT-24-25095; Revised: 01-Apr-2024, Manuscript No. AGT-24-25095 (R); Published: 08-Apr-2024, DOI: 10.35248/2168-9891.24.13.358
Abstract
Double-stranded RNA (dsRNA) triggers RNA interference (RNAi), leading to the directed silencing of specific genes. This mechanism has become significant and recognized as a sustainable tool in crop disease and pest- management applications. In vivo transcription of dsRNA in bacterial cells and in vitro transcription using kits are two methods of dsRNA production at laboratory scale. Design and testing of dsRNA at laboratory-scale requires efficient dsRNA production that yields large and high-quality dsRNA in a relatively lower price. In this report, we present an optimized protocol for production of double-stranded RNA (dsRNA) from bacterial cultures, achieving significantly higher yields compared to existing laboratory methods by five-fold (on average). Post extraction enzymatic digestion experiment revealed that the extracted dsRNA is pure and contains no trace amount of genomic DNA or single-stranded RNA (ssRNA) contamination. This study holds significant implications for RNAi-mediated gene silencing in agricultural biotechnology, particularly in crop protection.
Keywords
Agricultural biotechnology, Double-stranded RNA (dsRNA), RNA interference (RNAi), HT115 strain, Plant Pathogens
Introduction
RNA interference (RNAi) is a simple and rapid method of transcriptional as well as post-transcriptional gene silencing triggered by double-stranded RNA (dsRNA). This response leads to the reduction of expression and even suppression of specific genes either by breaking down their respective mRNA or by obstructing the translation process [1]. Ever since its initial identification in 1998 in the nematode Caenorhabditis elegans, RNAi has undergone significant development, evolving into a powerful molecular technique that can precisely inhibit gene expression [2]. It has extensive applications across various organisms. In addition to its fundamental role in gene function studies through RNAi- mediated gene silencing, RNAi is also being used for a wide range of practical applications. Spray Induced Gene Silencing (SIGS) technology is being developed as a potential bio-friendly alternative to the Virus Induced Gene Silencing (VIGS) and Host-Induced Gene Silencing (HIGS). For the field application of the exogenous dsRNA, either naked or nanoparticle mediated dsRNA can be applied onto the host species.
SIGS based RNAi technology has the potential to reduce the reliance on synthetic chemicals for controlling crop diseases and pests. Because of its potential, this technology has attracted the attention of environmentalists and investors in different companies such as Syngenta, Bayer as well as Greenlight Biosciences [3]. When proof of concept of the product is completed, a scale up and chemically producing the ribonucleic acids at large scale is economical. Such exogenous dsRNA treatments require 2-10 g of dsRNA per hectare [4]. This dosage may vary depending on the specific target organism, the crop, and other environmental factors. However, during R & D process, when the dsRNA molecules are being designed, produced, and tested at the laboratory scale, large scale production is not economical. R&D Laboratories uses either in vivo or cell-free methods (https://www.greenlightbiosciences.com, accessed on Jan 2, 2024) for the production of dsRNA to enable their design, development, and testing of their dsRNA.
Research on the exogenous application of the dsRNA for the plant disease management as well as gene function studies have been increased in the past years. For the laboratory scale, one way to do such studies is to use the commercially available kits such as Megascript kit or use of T7 RNA polymerase. Such options are highly expensive (Price for Megascript i~s 600 for 20 reactions) and thus therefore are not feasible for the repeated experiments. One of the best alternatives to such the expensive kits is microbial dsRNA production using bacteria in vivo. A key requirement for the success of RNAi applications is efficient production of dsRNA. Although, previous researchers have developed and proposed few protocols for the in vivo production and extraction of the dsRNA from bacterial cultures [5-9] improvement of dsRNA production and extraction protocol in a cost effective and reproducible manner is always in demand.
The in vivo synthesis strategy is more likely to reduce dsRNA production costs and increase yields in the future [10]. The dsRNA produced in engineered bacteria cannot be secreted directly outside the cell. Therefore, lysis, extraction and purification are required to obtain dsRNA. The lysis of cells can be performed by ultra-sonication, enzymatic lysis, boiling lysis, while Sodium Dodecyl Sulfate (SDS) can be used to enhance the lysis [11]. After the cell wall is broken, the nucleic acid can be released to obtain a crude extract of RNA. However, with the use of RNase A prior to TRIzol reagent pure dsRNA can be obtained. However, extracting, and purifying dsRNAs are relatively complicated and need to be further optimized.
Fusarium Head Blight (FHB) is a devastating fungal disease of hexaploid wheat (Triticum aestivum) caused by Fusarium graminearum. Availability of Resistant (R) genes against the Fusarium graminearum in wheat is limited to Fhb1 shown to provide a reasonably high level of genetic resistance against FHB, and lately Fhb7 which was shown to only decrease Deoxynivalenol (DON) content in grains post-infection [12-15]. This makes dsRNA induced gene silencing one of the alternative mode of resistance when R genes are rare such as Fusarium wilt in the tomato eggplant, bean, and watermelon.
This method is exactly what we adopted in our previous study in which the dsRNA production for RNAi was used for two genes- MGV1 and RAS1 in wheat against Fusarium graminearum [3]. Our previous study used the surface-functionalized graphene quantum dots (GQDs) as carriers to deliver double-stranded RNA (dsRNA) where the dsRNA-GQD conjugation was effective for the control of Fusarium graminearum. This method paper covers the detailed experimental approaches for five genes during the production of dsRNA for RNAi studies.
Materials and Methods
Gene selection and vector construction
This protocol was developed in the context of biocontrol of Fusarium head blight disease of wheat caused by fungus Fusarium graminearum. We selected genes that were previously shown to have a role in growth, development, Deoxynivalenol (DON) production and pathogenicity of the pathogen. These genes include MGV1, RAS1, YCK1, CAK1 and FgPp2A [16-19]. Firstly, we utilized the NCBI taxonomy browser to access the information on the Fusarium gramineraum strain PH-1. Subsequently, we navigated to the gene section and applied a filter by entering the gene ID to retrieve each of the gene sequences. The sequences were then subjected to a BLAST analysis against the Fusarium graminearum PH-1 (Taxid: 229533) genome database to identify potential homologous genes. For each gene, a 600-700 bp of the coding sequence was used to analyze and selection of the best regions for designing dsRNA. We analyzed the coding sequence by using the pssRNAit pipeline [20]. The search parameters applied to the pssRNAit were set to identify a 100-300 region with maximum numbers of predicted siRNA sites in Fusarium graminearum genome. The best predicted region by using pssRNAit pipeline, that is expected to yield several siRNA sites with high efficiency, was selected for cloning and in vivo transcription procedures. We further blasted the predicted regions for potential off-targets in the wheat genome as well as in human genome and confirmed no off- target sites in human and wheat.
In vivo transcription
Our approach for in vivo transcription involved cloning of the coding sequence of predicted region into an expression vector first. We used the plasmid vector, L4440- Addgene 1654. This vector was developed by Andrew Fire and Craig Mello as a revolutionary contribution to the field of RNA interference (RNAi) in the nematode Caenorhabditis elegans [2]. We used L4440 vector containing an ampicillin resistance gene and double T7 promoters in inverted orientation, flanking the multi-cloning sites [2]. For our design, we used restriction enzymes PstI and KpnI (New England Biolabs, MA). The recombinant L4440 vector harboring the coding sequence of predicted regions of each gene was transformed into the RNase III-deficient Escherichia coli strain HT115 (DE3) following the standard transformation procedure [21].
Induction of dsRNA
A single fresh colony of bacteria after transformation was inoculated into 3 mL LB+antibiotic (100 µg/mL ampicillin, 12.5 µg/mL tetracycline) and was grown to nearly stationary phase. Such culture was again inoculated to new LB media and was cultured overnight including antibiotics at 37 C with shaking overnight. Culture was further diluted to 1:100 with a final volume of 400 mL in LB+antibiotics and grown to OD600=0.4 with the shaking speed of 225 rpm. Such 400 ml was divided into four technical replicates each having 100 ml during the final dsRNA extraction. Bacterial cultures were induced to produce dsRNA by adding sterile IPTG to 0.5 mM [22]. After 2.5 hours, additional IPTG was added to a final concentration of 1 mM along with antibiotics (100 µg/mL AMP and 12.5 µg/mL/TET) and incubated at 37ºC with shaking additional 2.5 hours. Induced bacterial cultures were used for the purification of dsRNA following our optimized protocol.
Post-extraction enzymatic validation
This experiment was conducted to validate and confirm the absence of trace amount of genomic DNA or single-stranded RNA (ssRNA) in the extracted dsRNA. The dsYCK1 and dsFgPp2A extracts were subjected to DNase I (New England Biolabs) and RNase III (Thermo fisher scientific) treatment for further validation of product purity and quantification. With the treatment of DNase I, we aimed to remove any genomic DNA and with RNase III treatment, we aimed to confirm the presence of dsRNA in the samples. To do so, 7.5 microgram (μg) of each the dsYCK1 and dsFgPp2A were diluted in nuclease free water to a final volume of 25 μL for enzymatic treatments. Diluted samples were treated with either DNase I, RNase III, or both enzymes as per the manufactures’ instruction, and products were visualized with the samples receiving no enzymatic treatments on a 1.5 % agarose gel.
Reagents and equipment
• LB Broth Miller, fisher bioreagents, Lot: 225589
• Sodium dodecyl sulfate (Lauryl), Thermo Scientific, Ref: 28364, Lot: YB361182
• TRIzol Reagent, Invitrogen, Catalog number: 15596026
• Ethylenediamine tetraacetic acid (EDTA), Invitrogen by Thermo Scientific, PH 8.0, Lot: 01269799, Ref: AM9260G
• Tris-HCl, Invitrogen by Life Technologies, UltraPure 1M Tris- HCI, pH 8.0, Ref: 15568-025, Lot: 2517922
• Isopropyl-β-D-thiogalactopyranoside (IPTG) (Dioxane-free), Fisher BioReagents, Lot: 220900, CAS: 367-93-1
• RNase A, Thermo Scientific, REF: R1253
• Ampicillin Sodium Salt, Fisher BioReagentsTM
• Tetracycline, Thermo fisher scientific, Cas no: 60-54-8
• DNase I (RNase-free), New England Biolabs, Catalog#M0303S
• DNase I Reaction Buffer, New England Biolabs, Catalog#B0303S
• AmbionTM RNase III, Thermo fisher scientific, Catalog#AM2290 (10X Reaction Buffer included)
Buffer recipes
RNAse A buffer: Add 2.5 mL of 1M Tris-HCl and 1 mL of 0.5M EDTA, add autoclaved milli-Q water up to 50 mL.
Detailed extraction procedure
• Fill 50 ml of bacterial culture to the falcon tube, centrifuge@8000 RCF for 10 minutes@RT, discard the supernatant, and repeat the same procedure for the remaining 50 mL of culture.
• Add 5 mL of 0.25% SDS in the falcon tube either shake gently or use wide bore 5 mL pipette tips for the resuspension of the pellet.
• Boil the resuspended culture tube with SDS in 85°C for 5 minutes and incubate at RT for 10 minutes.
• Add 1.5 mL of RNAse buffer in each tube and add 10 ul of RNAse A solution in each tube.
• Incubate the solution in 37°C for 30 minutes. Again, incubate at 4°C for 10 minutes.
• Add 5 mL of TRIzol in each tube, mix and add 3 mL of chloroform, vortex gently and incubate at 4°C for 3 minutes.
• Centrifuge at 4°C for 15 minutes@8000 RCF.
• Pipet out upper layer (9 ml-10 ml) and add 6 mL of isopropanol and mix up and down for 4-6 times.
• Incubate at -20°C overnight, again centrifuge for 15 minutes at 4°C centrifuge@8000 RCF.
• Wash the pellet with 10 ml of 70% cold ethanol, centrifuge for 2 minutes at 4°C centrifuges@8000 RCF.
• Air dry the pellet for 30 minutes and re-suspend the pellet adding 10 mL of autoclaved ultrapure Milli-Q water, store in 4°C for short term storage and -20°C for long-term storage.
Results and Discussion
Production and validation of the dsRNA
This report presents an optimized protocol for production of dsRNA from bacterial cultures. The successful production of high-quality dsRNA across multiple and diverse sequences demonstrates the efficacy of the protocol. Extraction results of the dsRNA for five different dsRNA along with their quantity (µg of dsRNA/mL of bacterial culture) are presented in Figure 1. The quality parameters of all extracts for A260/280 ranged between 1.9 and 2.1 and the A260/230 parameters ranged between 1.7 to 2. Our results show the fast and straightforward production of dsRNA from the bacterial culture while maintaining higher yields without compromising the quality as well.

Figure 1: Concentration of dsRNA from bacterial culture for five genes of Fusarium graminearum per OD600 ml. The standard errors are based on four technical replicates each containing 100 mL of bacterial cultures. Note: Bacterial culture 
In enzymatic validation experiments performed for only two out of five genes, when dsRNAs were treated with DNase I, the results showed no trace amount of gDNA (Figure 2). However, when dsRNAs were treated with RNase III, a small sized fused band was seen for both genes, indicating the degradation of dsRNAs by the act of RNase III enzyme into the small RNA fragments producing small sized fused bands. RNase III family members bind with the dsRNA and cleave into two staggered sites eventually breaking the phosphodiester bonds of substrates and generate small sized fragments [23]. When both DNase I and RNase III were applied, fused bands were seen which were similar to the bands treated with RNase III only (Figure 2). Similar intensity and size of the RNA bands in the DNase I treated, and control samples indicates that no genomic DNA was present in the sample and the intense band represents the presence of the dsRNA.
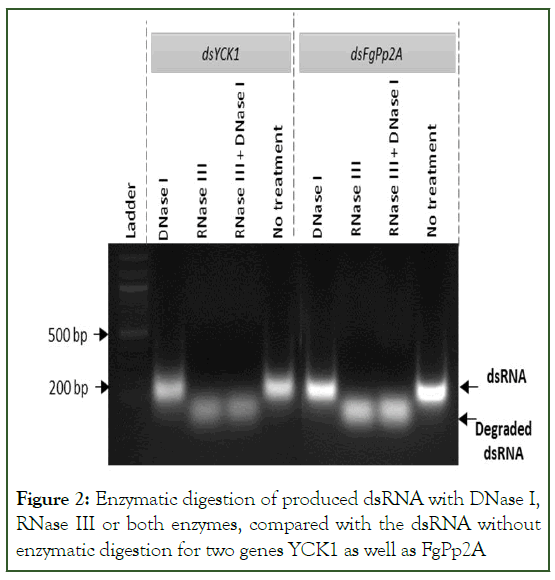
Figure 2: Enzymatic digestion of produced dsRNA with DNase I, RNase III or both enzymes, compared with the dsRNA without enzymatic digestion for two genes YCK1 as well as FgPp2A
Comparing dsRNA yields with existing methods
Using a review of the literature, we classified existing production protocols into three groups: conventional (phenol/chloroform) method heating method and sonication method [5-9,24-26]. The study by Ahn and colleagues shows that the estimated total dsRNA isolated by sonication was 488 µg per 25 mL culture (19.5 µg/mL), whereas heating and conventional were 239 µg per 25 mL culture (9.6 µg/mL) and 97 µg per 25 mL culture (3.9 µg/mL), respectively [7]. Another protocol resulted in significantly higher nucleic acid amounts as determined by spectrophotometric analysis (17.7 ± 1.24 µg/OD600 mL) following the modified heating and conventional method. One of the original studies on the dsRNA extraction by TRIzol method is a heating method and produced the 13.2 ± 1.00 µg/OD600 mL [5]. We used a modified SDS with heating method that resulted in production of approximately 63 µg/mL (averaged across five different dsRNAs in Figure 1) of the bacterial cultures, which is approximately five times greater than the average of productions in previous reports (Figure 3).

Figure 3: The concentrations of dsRNA produced and
reported by other studies in comparison to the yields obtained
in the current report. On average the yields in our report are 5-times greater than the average of earlier studies. Note: Bacterial culture 
The challenges and proposed modifications
Several methods have been devised for the extraction of dsRNA produced by bacteria, intended for applications in various research areas such as crop protection, vector control, and functional genomic analysis [5,7,11,27]. These protocols typically rely on commercially available phenol–guanidine-based reagents, such as TRIzol Reagent, which provide a comprehensive solution encompassing cell lysis, protein denaturation, and theinactivation of nucleases using the chaotropic agent guanidinium thiocyanate, combined with phenol. Among these methods, the protocol established by Ongvarrasopone and colleagues has consistently demonstrated superior results for dsRNA yield and purity [5]. A recent study has proposed an alternative approach by substituting TRIzolTM with a more cost- effective chemical mixture, P/C/I (25:24:1) at a slightly acidic pH range of 4.5-5. This alteration resulted in a higher incidence of co-purified bacterial RNAs and genomic DNA [28]. This situation necessities for the continued use of the TRIzol based dsRNA extraction protocol in laboratory scale although being slightly more expensive than the above-described chemical synthesis method.
Cell lysis represents an essential step in the process of extracting dsRNA produced by bacteria since failure to achieve proper lysis could result in the loss of dsRNA. Previous approaches have demonstrated that pretreating cells through sonication, heating, or enzymatic digestion can enhance the yield of dsRNA extraction. Nevertheless, it has been observed that dsRNA extracted from sonicated samples exhibits weaker bands and background smearing, indicating potential dsRNA degradation [7,25].
Our optimized dsRNA production protocol combines the factors from the conventional control and heating method with the use of TRIzol We have demonstrated that boiling bacterial cells in an optimized volume and concentration of the previously used SDS buffer effectively achieves cell lysis while maintaining both the quantity and quality of dsRNA (Figures 1 and 2) [5]. The use of 0.25% SDS and heat treatment at 85ºC for 5 minutes followed by incubation at room temperature is likely effective in lysing bacterial cells and denaturing proteins, facilitating the release of dsRNA. This eliminates the necessity for additional cell lysis treatments. In addition, high activity of RNase A can be obtained in the pH range of 7.6-8.0. Therefore, we changed the pH range to 8 instead of 7 (EDTA>PH 8.0, Tris-HCl>pH 8.0). The volume of TRIzol is one of the important factors in our extraction as deviation from that reduced the yield. Other modifications are incubation times and centrifugation parameters (see detailed extraction procedure section).
Significance of study
The significance of this study lies in facilitating laboratory scale dsRNA-based gene silencing, especially in the context of agricultural biotechnology and crop protection solutions. The ability to produce large quantities of high-quality dsRNA from bacterial cultures is a critical step in research and development phases of RNAi-based approaches for crop protection and trait development. The growing demand for pesticide-free crop production has increased the viability of using RNAi technology to address current agricultural and economic challenges [29]. Microbial dsRNA synthesis is generally considered as a potential strategy for reducing production costs in the future [10]. Use of dsRNA for the control of pathogens has already been successful in the fungal pathogens like Fusarium graminearum bacterial pathogens like Pectobacterium carotovorum, and viral pathogens like Tomato yellow leaf curl virus [3,30-32]. This approach can be extended to the other crops and pathogens including Fusarium wilt in the tomato eggplant, bean, and watermelon where numbers of R genes are limited and hard to deploy for providing the host resistance.
While dsRNA is environmentally safe and eco-friendly, the main challenge in utilizing it for agricultural purposes is the cost of large-scale production [33]. The expense associated with producing RNA presents a significant challenge to the widespread use of dsRNA spray technology. Current methods for dsRNA production include chemical synthesis, in vitro transcription and microbial fermentation. Chemical synthesis offers advantages such as producing a high yield of pure siRNA and a wider range of available modifications [34,35]. However, drawbacks include the cost and relatively lengthy turnaround times, typically ranging from 4 to 12 days, depending on synthesis and purification options. In Vitro Transcription (IVT) kits are more cost-effective compared to chemical synthesis, with the commercial MEGAscriptTM kit allowing the production of 1 gram for $3000 within a few hours [25]. Although in vitro transcription kits utilizing purified RNA polymerases and nucleotides have been widely used in laboratory experiments, but the high cost limits the large-scale application in the field [36,37].
Limitations of the study
Since RNA can be produced at very low cost by chemical synthesis, not all the researchers will have the access to the chemical synthesis, which is not also viable to produce for the small-scale laboratory studies. Large scale commercial application of the exogenous dsRNA cannot be reached by in vivo production systems using the large amounts of expensive chemical TRIzol, thereby also producing critical amounts of toxic organic waste. However, to make this economically viable, improvements in production and purification techniques are necessary to maximize dsRNA yield and quality in long run.
Additional considerations
• Extraction of the dsRNA on the same day after the completion of induction produces good yields and quality.
• RNA pellets should not be air-dried for more than one hour to prevent the possible degradation of dsRNA.
• Mixing and incubation with iso-propanol should not exceed 24 hours to prevent possible salt formation.
Conclusion
In this study, we present an optimized protocol for the laboratory scale dsRNA production from bacterial cultures using Fusarium graminearum genes as examples. Efficient production of dsRNA is a critical factor in the success of RNAi-based approaches, and our work addresses the need for cost-effective and high-yield methods for dsRNA production for such approaches. Our study explicitly provides a protocol to produce dsRNA with no contamination from the traces of genomic DNA as well as single stranded RNA. The significance of our work lies in its potential to enhance the feasibility of using RNAi-based approaches for crop protection, which aligns with the increasing demand for sustainable and pesticide-free agricultural practices, making our findings highly relevant to the agricultural biotechnology field.
Declarations
Funding
This research was funded by USWBSI agreements 59-0206-1-204 (Biodegradable nanomaterial-based non-GMO RNAi delivery for controlling FHB disease) and 59-0206-0-138 (Developing Native and Induce FHB Resistance Solutions in Wheat) generously made to MM. Supports from USDA Hatch grant 1013073 via Purdue College of Agriculture is appreciated.
Author contributions
Conceptualization, MM and HA; methodology, MM, HA, BG; formal analysis and investigation, BG; resources, MM; data curation, BG; writing-original draft preparation, BG and MM; writing-review and editing, MM; HA; supervision, MM; funding acquisition, MM; All authors have read and agreed to the published version of the manuscript.
Conflict of interest statement
Authors have no conflict of interest to report
References
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217-39.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nat. 1998;391(6669):806-811.
- Gyawali B, Rahimi R, Alizadeh H, Mohammadi M. Graphene Quantum Dots (GQD)-mediated dsrna delivery for the control of fusarium head blight disease in wheat. ACS Appl Bio Mater. 2024;7(3):1526-1535.
- Zotti M, Dos Santos EA, Cagliari D, Christiaens O, Taning CN, Smagghe G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag Sci. 2018;74(6):1239-50.
- Ongvarrasopone C, Roshorm Y, Panyim S. A simple and cost effective method to generate dsRNA for RNAi studies in invertebrates. Sci Asia. 2007;33(1):35-39.
- Okada R, Kiyota E, Moriyama H, Fukuhara T, Natsuaki T. A simple and rapid method to purify viral dsRNA from plant and fungal tissue. J Gen Plant Pathol. 2015;81:103-107.
- Ahn SJ, Donahue K, Koh Y, Martin RR, Choi MY. Microbial-based double-stranded RNA production to develop cost-effective RNA interference application for insect pest management. Int J Insect Sci. 2019;11:1179543319840323.
- Samarskaya VO, Spechenkova N, Markin N, Suprunova TP, Zavriev SK, Love AJ, et al. Impact of exogenous application of potato virus Y-specific DsRNA on RNA interference, pattern-triggered immunity and poly (ADP-Ribose) metabolism. Int J Mol Sci. 2022;23(14):7915.
- Ma Z, Zhang Y, Li M, Chao Z, Du X, Yan S, et al. A first greenhouse application of bacteria-expressed and nanocarrier-delivered RNA pesticide for Myzus persicae control. J Pest Scie. 2023;96(1):181-193.
- Cooper AM, Song H, Yu Z, Biondi M, Bai J, Shi X, et al. Comparison of strategies for enhancing RNA interference efficiency in Ostrinia nubilalis. Pest Manag Sci. 2021;77(2):635-645.
- Posiri P, Ongvarrasopone C, Panyim S. A simple one-step method for producing dsRNA from E. coli to inhibit shrimp virus replication. J Virol Methods. 2013;188(1-2):64-69.
- Jin F, Zhang D, Bockus W, Baenziger PS, Carver B, Bai G. Fusarium head blight resistance in US winter wheat cultivars and elite breeding lines. Crop Sci. 2013;53(5):2006-2013.
- Anderson JA, Glover K, Mergoum M. Successful adoption of spring wheat cultivars with moderate resistance to FHB by growers in the North Central Region. Proc. Natl. 2011.
- Gaire R, Brown-Guedira G, Dong Y, Ohm H, Mohammadi M. Genome-wide association studies for Fusarium head blight resistance and its trade-off with grain yield in soft red winter wheat. Plant Dis. 2021;105(9):2435-2444.
- Gyawali B, Scofield SR, Mohammadi M. Marker development and pyramiding of fhb1 and fhb7 for enhanced resistance to fusarium head blight in soft red winter wheat. Crops. 2023;3(4):320-332.
- Hou Z, Xue C, Peng Y, Katan T, Kistler HC, Xu JR. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol Plant-Microbe Interact. 2002;15(11):1119-1127.
- Bluhm BH, Zhao X, Flaherty JE, Xu JR, Dunkle LD. RAS2 regulates growth and pathogenesis in Fusarium graminearum. Mol Plant-Microbe Interact. 2007;20(6):627-636.
- Wang C, Zhang S, Hou R, Zhao Z, Zheng Q, Xu Q, et al. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 2011;7(12):e1002460.
- Yu F, Gu Q, Yun Y, Yin Y, Xu JR, Shim WB, et al. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014;203(1):219-32.
- Ahmed F, Senthil-Kumar M, Dai X, Ramu VS, Lee S, Mysore KS, et al. pssRNAit: a web server for designing effective and specific plant siRNAs with genome-wide off-target assessment. Plant Physiol. 2020;184(1):65-81.
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263(1-2):103-112.
- Teich A, Lin HY, Andersson L, Meyer S, Neubauer P. Amplification of ColE1 related plasmids in recombinant cultures of Escherichia coli after IPTG induction. J Biotech. 1998;64(2-3):197-210.
- Nicholson AW. Ribonuclease III mechanisms of double‐stranded RNA cleavage. Wiley Interdiscip Rev RNA. 2014;5(1):31-48.
- Chakraborty P, Ghosh A. Topical spray of dsRNA induces mortality and inhibits chili leaf curl virus transmission by Bemisia tabaci Asia II 1. Cells. 2022;11(5):833.
- Verdonckt TW, Vanden Broeck J. Methods for the cost-effective production of bacteria-derived double-stranded RNA for in vitro knockdown studies. Front Physiol. 2022;13:836106.
- Nerva L, Guaschino M, Pagliarani C, De Rosso M, Lovisolo C, Chitarra W. Spray‐induced gene silencing targeting a glutathione S‐transferase gene improves resilience to drought in grapevine. Plant Cell Environ. 2022;45(2):347-361.
- Solis CF, Santi-Rocca J, Perdomo D, Weber C, Guillen N. Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression. PLOS One. 2009;4(12):e8424.
- Figueiredo Prates LH, Merlau M, Rühl-Teichner J, Schetelig MF, Häcker I. An Optimized/Scale Up-Ready Protocol for Extraction of Bacterially Produced dsRNA at Good Yield and Low Costs. Int J Mol Sci. 2023;24(11):9266.
- Rank AP, Koch A. Lab-to-field transition of RNA spray applications–how far are we?. Front Plant Sci. 2021;12:755203.
- Koch A, Höfle L, Werner BT, Imani J, Schmidt A, Jelonek L, et al. SIGS vs HIGS: a study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non‐host plants. Molecular plant pathology. 2019;20(12):1636-1644.
- Mitter N, Worrall EA, Robinson KE, Li P, Jain RG, Taochy C, et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plant. 2017;3(2):1-0.
- Liu Y, Schiff M, Marathe R. Dinesh-Kumar SP. Tobacco Rattle Virus Mediates Gene Silencing in Plants by the Production of a Highly Effective Short Hairpin RNA. Plant Cell 2016; 28(1), 228-241
- Silver K, Cooper AM, Zhu KY. Strategies for enhancing the efficiency of RNA interference in insects. Pest Manag Sci. 2021;77(6):2645-2658.
- Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS letters. 2005;579(26):5974-5981.
- Tenllado F, Martínez-García B, Vargas M, Díaz-Ruíz JR. Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 2003;3:1-1.
- Rodrigues TB, Mishra SK, Sridharan K, Barnes ER, Alyokhin A, Tuttle R, et al. First sprayable double-stranded RNA-based biopesticide product targets proteasome subunit beta type-5 in Colorado potato beetle (Leptinotarsa decemlineata). Front Plant Sci. 2021;12:728652.
- He L, Huang Y, Tang X. RNAi-based pest control: Production, application and the fate of dsRNA. Front Bioeng Biotechnol. 2022;10:1080576.
Citation: Gyawali B, Alizadeh H, Mohammadi M (2024) An Optimized dsRNA Production Protocol from Bacterial Cultures: A Case for Fusarium graminearum Genes. Agrotechnology. 13:358.
Copyright: © 2024 Gyawali B, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


